INTRODUCTION
In recent years, the research based on the detection, impact, and degradation of antibiotics as micropollutants in environmental compartments, has notably increased (Gothwal and Shashidhar 2015, He et al. 2015). Recent reports described antibiotics input, occurrence, fate, and environmental effects (Sarmah et al. 2006, Kümmerer 2009, Gothwal and Shashidhar 2015). Contamination occurs due to the intensive use of antibiotics to treat both human and animal health, the wrong disposition, and inadequate treatment of effluents from the pharmaceutical industry or hospitals. Different classes of antibiotics have been detected, such as β-lactams, tetracyclines, aminoglycosides, macrolides, glycopeptides, quinolones, and sulfonamides, in water bodies and effluents of treatment plants (Fatta-Kassinos et al. 2011). Although these micropollutants are usually present at low levels, their adverse effects on aquatic life, animals, and humans are a growing global concern. One of the most claimed problems is the occurrence of antimicrobial resistance strains (Singer et al. 2016, Grenni et al. 2018).
Additionally, antibiotics can hamper the microbial community structure and function differently (Grenni et al. 2018). Due to their high dispersion in the environment, water treatment plants represent a crucial point for eliminating these pollutants. However, wastewater treatment plants (WWTP) are not entirely able to remove these trace organic compounds because they usually work to reduce the biochemical oxygen demand (BOD), nutrients, and microorganisms (He et al. 2015). Recent review articles on the methods for the degradation of antibiotics discuss the potentialities and limitations of such techniques and highlight the need for research before industrial application (Homem and Santos 2011, Méndez et al. 2017, Bairán et al. 2020). Among the proposed technologies, biocatalysis has emerged as an attractive alternative due to the already sustainable characteristics of the enzymatic processes (Wen et al. 2010, García-Zamora et al. 2018, Bilal et al. 2019). Enzymatic degradation of emergent micropollutants, such as hormones, anti-inflammatory drugs, antibiotics, and analgesic drugs, has been tested with peroxidases and laccases. The products of the enzymatic degradation by peroxidases and laccases are oxidized compounds, which are more biodegradable and with lower toxicity or biological activity than original micropollutants (Taboada-Puig et al. 2016, Zhang et al. 2016). In this work, we established the reaction conditions to oxidize sulfonamide antibiotics by versatile peroxidase (VP, EC. 1.11.1.16). VP is a heme peroxidase capable to oxidize several aromatic micropollutants since it contains a unique active site responsible for direct oxidation of various aromatic compounds; in addition, VP has an active site for manganese ion oxidation, which assists on the indirect oxidation of the same or other aromatic substrates.
MATERIALS AND METHODS
Chemicals
Sulfamethoxazole, sulfanilamide, sulfasalazine, sulfacetamide, sulfadiazine, hydrogen peroxide, and buffer salts were provided by Sigma-Aldrich (St. Louis, MO, USA). VP from Bjerkandera adusta UAMH 8258 was provided by Alltaenzymes (Edmonton, AB, Canada) with a Reinheitzahl ratio (Rz) (A403/A280) of 1.5 and a specific activity for manganese peroxidase (MnP) of 105 U/mg.
Enzymatic degradation of sulfonamides
The oxidation reaction of sulfonamide antibiotics was carried out at room temperature in a 1 mL reaction mixture (60 mM malonate buffer pH at 4.5, 10 µM antibiotics). The reaction conversion was estimated by monitoring the change in the substrate peaks over 10 min using a Perkin-Elmer HPLC system Series 200, equipped with a UV-vis detector and a binary pump. The concentration of the remaining substrate was measured every minute by stopping the reaction by adding 1 mL of 2-propanol. The peak areas were transformed into concentration values using a previously prepared standard curve for each antibiotic. The decrease in antibiotic concentration was plotted against time to determine the reaction constant, and the data were fitted to a first-order reaction using Origin 9.0 software (Originlab Corporation, Northampton, MA, USA). The reported values are the mean of three replicates.
Optimization of reaction conditions
A central composite design (CCD) with three independent variables (enzyme amount, reaction time, and hydrogen peroxide concentration at three levels) was applied to study the antibiotic conversion as the response pattern. The data were optimized using the Design-Expert software (v. 12 software trial, USA). Table I shows the experimental values for the variables. Twenty experiments were conducted according to the CCD.
TABLE I CENTRAL COMPOSITE DESIGN FOR SULFONAMIDE OXIDATION BY VERSATILE PEROXIDASE.
| Assay | H2O2 (mM) | Enzyme (U) | Time (min) | Conversion (%) |
| 18 | 1 | 3.2 | 5 | 32.58 |
| 9 | 0.67 | 1.76 | 10 | 43.79 |
| 6 | 0.33 | 3.2 | 15 | 96.57 |
| 13 | 0.67 | 3.2 | 10 | 65.48 |
| 2 | 0.67 | 1.76 | 10 | 52.73 |
| 10 | 1 | 1.76 | 10 | 49.25 |
| 20 | 0.67 | 1.76 | 10 | 44.99 |
| 5 | 0.67 | 1.76 | 10 | 38.42 |
| 7 | 0.33 | 0.32 | 15 | 11.81 |
| 4 | 0.33 | 0.32 | 5 | 9.87 |
| 1 | 1 | 0.32 | 15 | 8.32 |
| 15 | 0.33 | 1.76 | 10 | 64.23 |
| 19 | 0.67 | 1.76 | 15 | 59.28 |
| 8 | 0.67 | 1.76 | 5 | 45.98 |
| 16 | 1 | 3.2 | 15 | 56.56 |
| 3 | 0.67 | 1.76 | 10 | 59.79 |
| 14 | 0.33 | 3.2 | 5 | 60.47 |
| 17 | 0.67 | 0.32 | 10 | 13.19 |
| 12 | 1 | 0.32 | 5 | 7.59 |
| 11 | 0.67 | 1.76 | 10 | 54.08 |
Docking analysis
The crystallized structure of the dihydropteroate synthase (DHPS) from Escherichia coli was employed for this analysis (Protein Data Bank ID: 1ajz) (Achari et al. 1997). Three-dimensional structures of dihydropterin pyrophosphate (DHPPP), p-aminobenzoic acid (pABA), sulfasalazine, and sulfamethoxazole were obtained from commercially available compounds database ZINC12. The 3D structures belong to sulfasalazine-degradation products (Sz-degradation products 1 and 2) and sulfamethoxazole-degradation products (Sm-degradation product 1), which were obtained with the program ChemSketch. A geometrical optimization for each ligand was performed by using Avogadro software (Hanwell et al. 2012). The molecular mechanical (MM) calculation based on the Merk molecular force field and a steepest descent algorithm was employed for the optimization, allowing that all atoms were movable (see Fig. S1 in the supplementary material). Docking analysis was performed to elucidate the binding orientation of the ligands with DHPS by using Autodock 4 (Morris et al. 2009). Parameters for the simulation were established accordingly to a previously parametrization method (Andrade-Pavón et al. 2019, Ortiz-Álvarez et al. 2020). The establishment of the grid dimension was performed based on the location of amino acid residues previously mentioned by Yun et al. (2012). The grid dimensions for DHPS-DHPPP were 40 × 50 × 54 Å3 with points separated by 0.464 Å, and the following grid center: X = 40.075, Y = 4.776, and Z = 4.683. The grid dimensions for DHPS-pABA, DHPS-sulfa drugs, and DHPS with the degradation products derived from sulfa drugs (DPSD) were 48 × 52 × 40 Å3 with points separated by 0.375 Å, and the following grid center: X = 40.585, Y = 7.128, and Z = 4.993. A total of 100 runs by triplicate, computing with a maximum number of eval of 25 000 000 and by using a specifical number of generator seeds (DHPPP: 1624358890; pABA: 1627240940; sulfasalazine: 1626479527; sulfamethoxazole: 1626979527; sulfapyridine: 1626364935; 2-benzenesulfonylaminopyridine: 1626667424; 4-nitro sulfamethoxazole: 1626537416) was established for docking simulation. Results of docking were visualized employing the software LigProt+ (Laskowski and Swindells 2011).
High performance liquid chromatography (HPLC) analysis
The substrate concentrations were measured using a Perkin Elmer HPLC system (Hopkinton, MA, USA) equipped with a reversed-phase C-18 column (Genesis, 5 × 125 mm, 5 µm), a binary pump system, and a 20 µL injection loop, and they were eluted using a pH 2 acetonitrile-phosphate buffer solvent mixture at 0.7 mL/min. Substrate detection was carried out using UV-vis detection. The HPLC conditions used for separation and detection are listed in table II.
TABLE II LIQUID CHROMATOGRAFY (HPLC) CONDITIONS FOR THE TESTED SULFONAMIDE ANTIBIOTICS.
| Antibiotic | Mobile phase | Detection wavelength (nm) | Retention time (min) |
| Sulfamethoxazole | Acetonitrile and phosphoric acid 20 mM (30:70) | 270 | 7.19 |
| Sulfadiazine | Acetonitrile and phosphoric acid 20 mM (10:90) | 265 | 9.82 |
| Sulfanilamide | Acetonitrile and phosphoric acid 20 mM (10:90) | 259 | 4.12 |
| Sulfacetamide | Acetonitrile and phosphoric acid 20 mM (25:75) | 268 | 5.54 |
| Sulfasalazine | Acetonitrile and phosphoric acid 20 mM (70:30) | 350 | 2.92 |
All HPLC runs were done at pH 2.
Products identification
Reaction products were identified by LC-MS (Chromatograph Series 1260) coupled to ESI-Q-TOF-MS (6520, Agilent Technologies, Germany). The separation was done on a Zorbax Eclipse XDB-C18 Narrow-Bore column (2.1 × 150 mm, 5 µm) using the mobile phase acetonitrile-water both with 0.1 % HCOOH to promote ionization. Gradient elution was performed from 10-30 % of acetonitrile in 15 min, then back to 10 % in 3 min followed by long enough to stabilize the baseline, flow was maintained at 1 mL/min. The parameters of the ESI source were the following: positive ionization mode, voltage of fragmenter 175 V, capillary voltage 3500 V, gas temperature 350 ºC, N2 flow 11 L/min, nebulizer 60 psi. The flow velocity was 0.6 mL/min.
Assays in treated wastewater
The oxidative capability of VP was determined for all the sulfonamide antibiotics in real wastewater effluent from a secondary treatment outlet of a municipal treatment plant of the state of Puebla, Mexico. The wastewater sample was physical and chemically characterized according to standard methods, and its main parameters are summarized in table III. Water samples were spiked with 10 μM of each sulfonamide, and the pH was fitted to 4.5 using a solution of HCl 0.1 N. Three replicate experiments were performed for all samples.
RESULTS
Effect of the reaction conditions on the oxidation of sulfamethoxazole
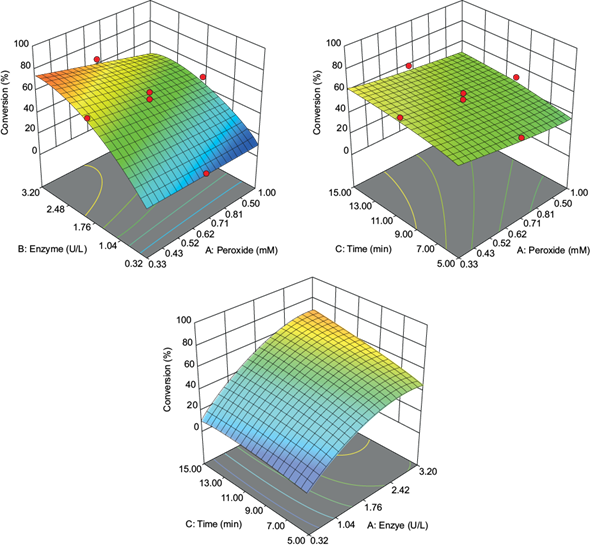
In the preliminary assessments (60 mM malonate buffer pH at 4.5, 10 µM antibiotic, 3.2 U VP, 1 mM H2O2, 10 min reaction time), the enzyme was not capable of oxidizing the sulfonamide antibiotics in the presence of manganese sulfate. However, the manganese ion was easily oxidized (standard catalytic activity of 105 U/mg.). Previously, Eibes et al. (2011) reported a high rate of conversion for the oxidation of sulfamethoxazole using VP in the presence of Mn2+ ions. The difference in the biocatalytic performance between that study and this work may be due to the longer reaction time (7 h) that would allow for substrate oxidation (Eibes et al. 2011). In the absence of manganese ions, the enzyme oxidized all sulfonamide antibiotics to different extents. The optimization of reaction conditions was carried out using sulfamethoxazole as a model substrate for the CCD assays (Table I). As can be seen, the variables had a pronounced effect on the enzyme’s transformation capacity, with conversion values from 7 to 96 %. The relationship between variables and conversion was adjusted to the second-order statistical model:
where t = time, E = enzyme.
The model and its terms were significant at the 5 % level (Table III). The most relevant independent variable was the enzyme amount, followed by the hydrogen peroxide concentration. Time did not show a significant effect (at least for the interval assayed as the reaction is finished in a shorter reaction time, see below). Based on this model, response surface graphs were constructed, showing that a high enzyme concentration, prolonged reaction time, and low hydrogen peroxide concentration resulted in the highest conversion rates (Fig. 1). The obtained optimal values, using a desirability value of 1, were 0.33 mM of hydrogen peroxide, 3.2 U of the enzyme, and a 15 min reaction time. Like other peroxidases, excess hydrogen peroxide inactivates VP; so, an adequate amount of H2O2 is relevant for suitable enzyme stability and high antibiotic conversion. Reaction times set for 15 min achieved conversions more elevated than 90 %. This reaction time is similar to those shown by other peroxidases, and shorter than those showed by laccases for the oxidization of pharmaceutical micropollutants (Weng et al. 2012, Becker et al. 2016, Zhang et al. 2016).
TABLE III ANALYSIS OF VARIANCE (ANOVA) FOR THE CONVERSION EQUATION.
| Source | Sum of source | df | Mean squares | F value | p-value prob > F |
| Model significant | 8727.33 | 9 | 969.70 | 21.27 | < 0.0001 |
| A-peroxide | 520.80 | 1 | 520.80 | 11.42 | 0.0070 |
| B-enzyme | 5981.72 | 1 | 5981.72 | 131.22 | < 0.0001 |
| C-time | 354.89 | 1 | 354.89 | 7.78 | 0.0191 |
| AB | 259.90 | 1 | 259.90 | 5.70 | 0.0381 |
| AC | 1.25 | 1 | 1.25 | 0.028 | 0.8715 |
| BC | 209.55 | 1 | 209.55 | 4.60 | 0.0576 |
| A2 | 5.01 | 1 | 5.01 | 0.11 | 0.7471 |
| B2 | 699.12 | 1 | 699.12 | 15.34 | 0.0029 |
| C2 | 19.26 | 1 | 19.26 | 0.42 | 0.5304 |
| Residual | 455.87 | 10 | 45.59 | ||
| Lack of fit | 144.56 | 5 | 28.91 | 0.46 | 0.7901 |
| Not significant | |||||
| Pure error | 311.31 | 5 | 62.26 | ||
| Cor total | 9183.20 | 19 |
Under optimal reaction conditions, the attained conversion percentages for sulfanilamide, sulfadiazine, sulfacetamide, and sulfasalazine were 94, 100, 95, and 100 %, respectively.
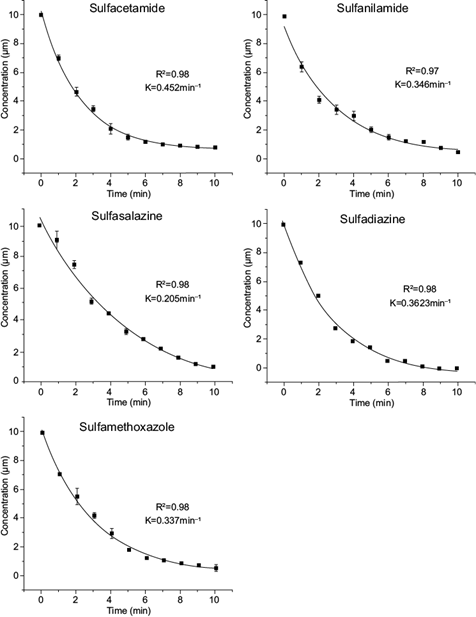
Because the concentration of antibiotics as contaminants is very low (μg/L or ng/L), the kinetic behavior of the enzyme is usually due to a first-order mechanism, which indicates that the reaction rate is linearly proportional to the concentration of the substrate. The reaction rate constant (k) for the oxidation of the five sulfonamides by the VP was determined. The decline of all sulfonamides was consistent with a first-order kinetic model (Fig. 2). The conversion and k values shown by VP were higher for the transformation of other sulfonamides catalyzed by horseradish peroxidase and soybean peroxidase (al-Maqdi et al. 2018, Yang et al. 2018). The k values provided t1/2 values similar to those for the oxidation of the antibiotics sulfadimethoxine and sulfamonomethoxine by laccase, using violuric acid and 2-2’-azinobis (3-ethylbenzthiazoline-6-sulfonic acid) as redox mediators (from 1.5 to 3.38 min) (Weng et al. 2012).
Identification of reaction products
The identification of the enzymatic reaction products of sulfonamides has been reported for peroxidases and laccase, although there has been no clear identification of the oxidized products by the latter (Eibes et al. 2011, Weng et al. 2012, Zhang et al. 2016). In this study, only the reaction products of sulfasalazine and sulfamethoxazole could be identified (Table IV, Fig. 3). For the rest of the compounds, we observed insoluble precipitates that were not analyzed. The products identified for sulfasalazine oxidation, 2-benzenesulfonylaminopyridine, and sulfapyridine, were also determined before by a Fenton-like reaction (Fan et al. 2011). For sulfamethoxazole, oxidation transformed the amino group into the nitro group, and the same product, WWTPsulfamethoxazole, was identified for the oxidation using potassium ferrate combined with ultrasound (Zhang et al. 2015). Other reaction products have been reported for peroxidases enzymes such as soybean peroxidase, chloroperoxidase, and versatile peroxidase in the oxidation of sulfamethoxazole, the reported products exhibited lower molecular weights, indicating that a hydrolytic reaction took place after the enzymatic oxidation (Eibes et al. 2011, Zhang et al. 2016, al-Maqdi et al. 2018); however, in this report, none of these products were found. More studies are needed to examine the oxidation pathway carried out for versatile peroxidase.
TABLE IV IDENTIFIED REACTION PRODUCTS FROM SULFONAMIDE ANTIBIOTICS OXIDATION BY VERSATILE PEROXIDASE.
| Compound | Experimental m/z [M+H]+ | Theoretical m/z [M+H]+ | Error (ppm) | Retention time (min) | Reference |
Sulfasalazine (PM 398.39) 
|
399.0789 | 399.0758 | 7.87 | 14.451 | |
Reaction product 1 (2-benzenesulfonylaminopyridine) 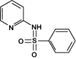
|
235.0506 | 235.0536 | 0.11 | 6.775 | (Fan et al. 2011) |
Reaction product 2 (Sulfapyridine) 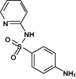
|
250.0633 | 250.0645 | 4.71 | 11.28 | (Fan et al. 2011, Saini and Bansal 2014) |
Sulfamethoxazole (PM 253.279) 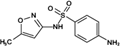
|
254.0680 | 254.0594 | 5.58 | 5.876 | |
Reaction product (WWTP sulfamethoxazole) 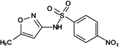
|
284.0343 | 284.0336 | 2.59 | 14.081 | (Zhang et al. 2015) |
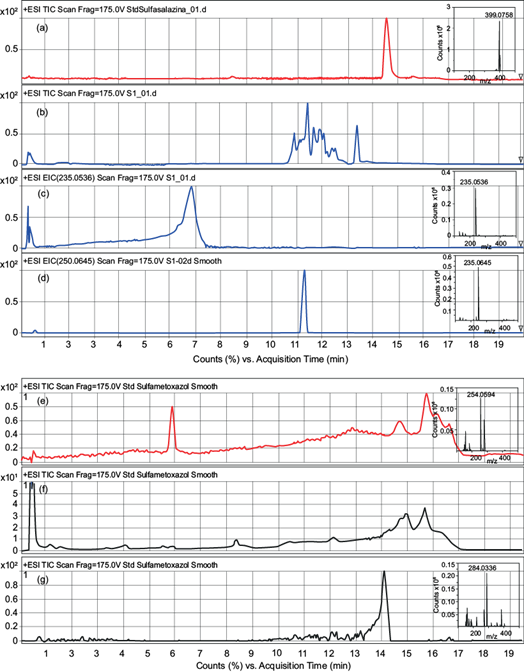
Fig. 3 Chromatograms of compounds reported in table IV and their respective mass spectra identified in the inset. (a) Total ion chromatogram (TIC) of sulfasalazine standard [M+H]+=399; (b) TIC of sulfasalazine solution after enzymatic degradation and the respective (c) extracted ion chromatogram (EIC) of the mass [M+H]+=235 and (d) mass [M+H]+=250; (e) TIC of sulfamethoxazole standard [M+H]+=254; (f) TIC of sulfamethoxazole solution after enzymatic degradation and the respective (g) EIC of the mass [M+H]+=284.
Based on the identification of the reaction products generated with the biocatalytic treatment, the antibacterial activity of these products can be assessed based on previous reports. Greenstein et al. (2007) evaluated the antimicrobial activity of sulfapyridine in Mycobacterium avium subspecies paratuberculosis, a bacterium of veterinary importance. The inhibitory tests showed that M. avium ssp. showed a high susceptibility to sulfapyridine, and sulfasalazine does not have a pronounced effect. On the other side, Andreasen et al. (1988), reported the minimum inhibitory concentration (MIC) values for sulfasalazine and sulfapyridine against gram-negative enteric bacteria. All strains assayed were resistant to 1600 mg/mL of sulfasalazine; meanwhile, for sulfapyridine, the MIC range for Yersinia enterocolitica was 3.1-25 mg/mL and for Salmonella 25-100 mg/mL. Shigella and three of five E. coli strains were resistant to 1600 mg/mL of sulfapyridine. Two strains of E. coli were inhibited by 25 mg/mL. Both reports show that the antibacterial activity is higher than the parental compound sulfasalazine.
Igwe and Okoro (2014) evaluated the antimicrobial activity of 2-benzenesulfonyl-aminopyridine against eight microorganisms, six bacteria (Bacillus cereus, Sarcina lutea, Pseudomonas aeruginosa, Escherichia coli, Salmonella typhi, Klebsiella pneumoniae) and two fungi (Candida albicans and Aspergillus niger). The results showed that 2-benzenesulfonyl-aminopridine has antimicrobial effect against B. cereus, S. lutea, S. typhi, C. albicans and A. niger.
Finally, Majewsky et al. (2014) evaluated the antimicrobial activity of WWTPsulfamethoxazole, among other 11 transformation products of sulfamethoxazole on Vibrio fischeri. The minimum inhibitory concentration results showed that WWTP sulfamethoxazole had better antimicrobial activity against V. fischeri than sulfamethoxazole.
Since not all the reaction products or intermediaries were identified, it is necessary to carry out further studies to define the degradation pathway; in addition, the possible remaining toxicity of the products should be determined.
In silico binding of sulfonamide reaction products with DHPS
Docking analyses displayed a hypothetical binding mode of substrates, sulfa drugs and DPSD with the catalytic site of DHPS. The binding energy values among ligands and DHPS are summarized in table V. A natural substrates of DHPS, pABA, was bound with a binding energy of 5.69 kcal/mol, while sulfa drugs and DPSD displayed higher binding energy than pABA (Table V). Sulfapyridine and 2-benzenesulfonylaminopyridine bound with lower affinity than sulfasalazine, but WWTP sulfamethoxazole displayed higher affinity upon the catalytic site of the DHPS than sulfamethoxazole (Table V).
TABLE V DOCKING RESULTS OF SULFONAMIDE COMPOUNDS AND THEIR DERIVED DEGRADATION PRODUCTS WITH DIHIDROPTEROATO SINTASA.
| Compound | Binding energy (kcal/mol) |
| 6-hydroxymethyl-7, 8-dihydropterin pyrophosphate | -8.54 ± 0.0034 |
| p-aminobenzoic acid | -5.69 ± 0.052 |
| Sulfasalazine | -7.95 ± 0.045 |
| Sulfapyridine | -6.94 ± 0.017 |
| 2-benzenesulfonylaminopyridine | -6.47 ± 0.051 |
| Sulfamethoxazole | -7.24 ± 0.26 |
| 4-nitro sulfamethoxazole | -8.09 ± 0.22 |
Figure 4 describes the interactions among ligands and amino acid residues located in DHPS. pABA exhibited hydrogen bonds with a length of 2.64-3.04 Å between Asn22, Arg63, Glu60, and Asp96 in the carboxyl group at C-1 and amino group located at C-5 (Fig. 4a). Sulfasalzine (Fig. 4b) was capable of interacting with Asp56, Thr62, Arg63, Lys221, Ser222, and Arg235, with a length hydrogen bonds of 2.79 -3.32 Å, binding it with the carboxyl group located at C-15, the oxygen and nitrogen atoms of the sulfa group located at N1 and O1. Sulfamethoxazole (Fig. 4c) interacted only with Glu60 and Arg63, but short bond lengths of 2.50-2.75 Å were observed in the amino group located at C-8 and O1 and O2 of the sulfa groups. Sulfapyridine (Fig. 4d) displayed hydrogen bonds between Thr62 with the oxygen O1 of the sufonyl Asp96 and the nitrogen N2 of pyridine group. The 2-benzenesulfonylaminopyridine (Fig. 4e), bound with Thr62 at O1 of the sulfa group and the amino group at C9 with Gly217, displaying bond lengths between and 2.79-3.10 Å. Finally, WWTP sulfamethoxazole (Fig. 4f) showed interactions at O4 and O5 of the nitro group with Asp185, Phe188, and Gly217, with bounds lengths of 2.60-2.83 Å.
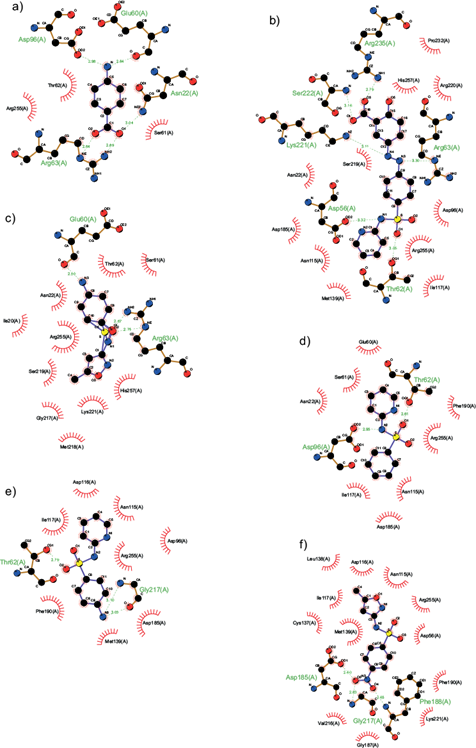
Fig. 4 Predicted binding mode of (a) p-aminobenzoic acid (pABA), (b) sulfasalazine, (c) sulfamethoxazole, (d) 2-benzenesulfonylaminopyridine, (e) sulfapyridine, and (f) WWTP sulfamethoxazole with the catalytic site of dihydropteroate synthase. The docking analyses were performed with Autodock 4. In the 2D model obtained with the LigProt+ software, electrostatic van Deer Waals interactions among amino acid residues and the ligands are portrayed as red rays. Hydrogen bonds are depicted by green dotted lines and the distances of these bonds are denoted in Angstroms.
Degradation efficiency achieved using the VP/H2O2 system to treat sulfonamides in residual-treated wastewater
The capability of VP to oxidize sulfonamide antibiotics was determined in samples of residual treated wastewater taken from the wastewater treatment plant of San Simón Atzitzintla, located in the municipality of San Salvador el Verde, state of Puebla (Table VI). These water samples did not have measurable quantities of the tested sulfonamides, as assayed by HPLC-MS; however, a high chemical oxygen demand (COD) value and low BOD value were found, indicating the presence of recalcitrant compounds in the effluent. In addition, high contents of nitrates and phosphates were determined, suggesting a low efficiency of the treatment plant for nutrient removal. Under the optimal reaction conditions, the attained enzymatic conversions were higher than 85 % for sulfamethoxazole, sulfasalazine, and sulfadiazine. Meanwhile, lower conversion percentages were reached for sulfacetamide (47 %) and sulfanilamide (13 %). The lower conversion percentages in the wastewater-treated effluent could be due to natural organic matter, as it is already known that the organic matter content has an inhibiting effect on several conversion oxidative technologies (Gao et al. 2014).
TABLE VI PHYSICOCHEMICAL PROPERTIES OF RESIDUAL TREATED WASTEWATER.
| Parameter | Nom | Maximum allowable limit | Value obtained |
| Fe+2 | NOM-127-SSA1-1994 | 0.30 mg/L | 0.178 mg/L |
| SO4 -2 | NOM-127-SSA1-1994 | 400 mg/L | 90 mg/L |
| P | * | * | 14.4 mg/L |
| P2O5 | * | * | 31 mg/L |
| PO4 -3 | * | * | 40.2 mg/L |
| NO3-N | NOM-127-SSA1-1994 | 10.00 mg/L | 24 mg/L |
| NO3 -1 | NOM-127-SSA1-1994 | 0.05 mg/L | 104 mg/L |
| Free chlorine | NOM-127-SSA1-1994 | 0.2-1.50 mg/L | 0.1 mg/L |
| Mg+2 | * | * | 10 mg/L |
| Ca+2 | * | * | 145 mg/L |
| Biochemical oxygen demand | NOM-003-ECOL-1997 | 20 mg O2/L direct contact 30 mgO2/L indirect or casual contact | 2.05 mg O2/L |
| Chemical oxygen demand | * | * | 651.38 mg O2/L |
| Conductivity | * | * | 1448 µs/cm |
| pH | * | 6.5-8.5 | 7.0 |
*There is no offcial regulation for this parameter.
DISCUSSION
The application of enzymes in environmental treatment is becoming a promising technology due to the managed reaction conditions such as low energy demand, zero sludge generation, high pollutant conversion percentages, and low product toxicity of the reaction. Some enzymes have been applied in biocatalytic formulations immobilized in membranes and tested in reactors under real conditions found in WWTP with promising results (Longoria et al. 2010, Touahar et al. 2014). Peroxidases and laccases have been proposed and tested against an important battery of microcontaminants, resulting in oxidized compounds with higher biodegradability. However, as in this and other reported cases, the product toxicity should be evaluated before implementing the biocatalytic technology to avoid the production of more toxic compounds (Becker et al. 2016). Taken together, the results of this study demonstrate VP can transform sulfonamide micropollutants at high velocity and conversion rates; however, the generation of products with higher antimicrobial activity is a challenging issue that must be resolved before proposing the enzyme as part of an eventual polishing technology in wastewater treatment facilities.
In the reaction mechanism of the enzyme DHPS which is the target of sulfonamide antibiotics, a main molecular event is the interaction of DHPS with the free amino group NH2 in position 4 of its natural substrate (pABA) in a condensation reaction. A great concern is to know whether DPSD might exhibit affinity upon the catalytic pocket of DHPS, consequently resulting in the promoting of microbial resistance. For obtaining a rough response, docking simulation was carried out by using DHPS from E. coli, which is a microorganism with a high prevalence in wastewaters and possesses resistance to sulfonamides (Bean et al. 2009, Gekenidis et al. 2018).
According to the results summarized in table V, DPSD derived from sulfasalazine exhibited a low docked energy in interaction with the catalytic portion of DHPS, compared to the results obtained with sulfasalazine. Conversely, the degradation products showed more affinity in comparison with pABA. This could be explained by the fact that DPSD derived from sulfasalazine displayed a higher number of interactions than the original substrate (pABA) upon DAPH, while pABA is capable of binding to catalytic site with only seven amino acid residues (Fig. 4). On the other hand, the DPSD displayed several hydrogen bonds, perhaps more complex in their chemical structure. The results show that the presence of a benzenesulfonamide group provides a better affinity enhancing the interaction in sulfa drugs and their DPSD due to a significant number of contacts and shorter hydrogen bonds (Fig. 4). The DPSD derived from sulfamethoxazole showed a higher docked energy than the results obtained with sulfamethoxazole (Table V).
CONCLUSION
Versatile peroxidase seems to be a potential catalyst for the oxidization of sulfonamide antibiotics in real treated wastewater effluents. Biocatalysis could be applied as a polishing technology to oxidize persistent antibiotic micropollutants only if there is a complete assurance that no more toxic products are generated. However, docking analyses suggest that degradation products may still promote the appearance of drug resistance in bacteria; therefore, it is necessary to find strategies for enhancing the capability of the peroxidase to degrade the antibiotics completely. In this work, the identified products have higher antibacterial activity; therefore, it is necessary to deepen the studies to identify the rest of the reaction products, elucidate the degradation mechanism and determine the toxicity and biodegradability of the reaction products.











 nova página do texto(beta)
nova página do texto(beta)