Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista internacional de contaminación ambiental
versão impressa ISSN 0188-4999
Rev. Int. Contam. Ambient vol.25 no.3 Ciudad de México Ago. 2009
Bacterial population dynamics and separation of active degraders by stable isotope probing during benzene degradation in a BTEX–impacted aquifer
Dinámica de poblaciones bacterianas y separación de degradadores activos por medio de isótopos estables durante la degradación de benceno en un acuífero contaminado con BTEX
Arturo ABURTO1,2,*, and Andrew S. BALL1,3
1 Department of Biological Sciences, University of Essex, Wivenhoe Park, Colchester CO4 3SQ, United Kingdom.
2 Present address: Universidad Autonoma Metropolitana, Artificios 40, Miguel Hidalgo, Cuajimalpa, México.
3 Present address: School of Biological Sciences, Flinders University of South Australia, GPO Box 2100, Adelaide, South Australia 5001.
* Corresponding author:
Arturo Aburto.
Tel.: +52 55 58044600 ext 2677,
Fax: +52 55 58046407.
E–mail address: aaburto@correo.cua.uam.mx
Recibido agosto 2008
Aceptado febrero 2009
ABSTRACT
The activity and diversity of a groundwater bacterial community was studied during the degradation of benzene in samples from a BTEX–contaminated aquifer (SIReN, UK) through the use of denaturing gradient gel electrophoresis (DGGE), followed by excision and sequencing of dominant bands. Rapid aerobic benzene degradation occurred in all samples, with 60–70 % degradation of benzene. DGGE analysis revealed that unique, stable bacterial communities were formed in each sample. Pseudomonas putida and Acidovorax delafieldii were identified in groundwater samples 308s and W6s respectively, suggesting they are the important taxa involved in the degradation of benzene. Further work based on stable isotope probing (SIP) of RNA using 13C benzene was carried out. Prominent bands were identified as Acidovorax and Malikia genera; the latter is very similar to the benzene–degrader Hydrogenophaga, which confirms the presence of active benzene degraders in the groundwater samples. The identification of the prominent communities provides knowledge of the bioremediation processes occurring in situ and the potential to enhance degradation. This study highlights the potential of combining community fingerprinting techniques, such as DGGE, together with SIP.
Key words: aerobic benzene degradation, SIP, groundwater, population dynamics.
RESUMEN
El presente estudio investiga la dinámica de poblaciones durante la degradación de benceno en muestras de un acuífero contaminado con BTEX mediante la técnica de DGGE, obtención de bandas y su posterior secuenciación. Los resultados indican degradación aeróbica de benceno en todas las muestras y que las poblaciones son diferentes en cada muestra, pero se mantienen estables durante la degradación del contaminante. Las principales bandas obtenidas en el gel de poliacrilamida fueron secuenciadas e identificadas como Pseudomonas putida y Acidovorax delafieldii en las muestras 308s y W6s respectivamente, lo que sugiere que estos microorganismos son los principales degradadores en ciertas muestras. Mediante el uso de isótopos estables se comprobó la existencia de microorganismos aerobios degradadores de benceno, los cuales fueron identificados como miembros de los géneros Acidovorax y presumiblemente Hydrogenophaga en otra de las muestras. La identificación de comunidades prominentes brinda información sobre los procesos que ocurren in situ y del potencial para aumentar la degradación. El presente estudio muestra el potencial de combinar técnicas como DGGE con isótopos estables.
Palabras clave: degradación aeróbica de benceno, SIP, agua subterránea, dinámica de poblaciones.
INTRODUCTION
The contamination of groundwater represents a major hazard in terms of both environmental damage and public health; contaminants such as PAH (polyaromatic hydrocarbons), MTBE (methyl tert–butyl ether), and BTEX (benzene, toluene, ethylbenzene and xylenes) enter natural waters via wastewater effluents from petroleum refining industries, rainwater runoff from roads, accidental spills and leakages from underground storage tanks. The most soluble components of gasoline and diesel fuel are the BTEX compounds, making them common contaminants in a large number of aquifers. Among the BTEX compounds, benzene requires special attention because it is the most toxic, the most soluble (Sikkema et al. 1995) and carcinogenic (Dean 1985). Benzene is readily degraded aerobically, and recent studies have shown it is also degraded in the absence of oxygen (Phelps et al. 1998, Rooney–Varga et al. 1999, Caldwell and Suflita 2000, Villatoro–Monzón et al. 2003, Kasai et al. 2006). Further work is now required to elucidate the microorganisms responsible for benzene degradation in the various environments.
Stable isotope probing
Stable isotope probing (SIP) is used to identify microorganisms that assimilate a specific growth substrate. This is achieved by labelling the substrate with a stable isotope (the most common is 13C) that will be incorporated into the microorganism's cellular biomarkers (e.g. lipids, DNA and rRNA) if they utilize the substrate. The labelled nucleic acids can then be resolved from the unlabelled ones by density gradient ultracentrifugation, specific genes amplified and separated in a denaturing gel, and sequenced. Thus, by sequencing the labelled nucleic acids, the microorganisms responsible for the substrate uptake can be identified and therefore link function to microbial identity. Although SIP can be performed with DNA or rRNA, the latter presents some advantages over DNA, such as a higher copy number and a higher turnover rate, which is a reflection of cellular activity independent of replication (Manefield et al. 2002a, Manefield et al. 2002b, Lueders et al. 2004, Dumont and Murrell 2005).
The aim of this study was to obtain a profile of the bacterial community in several groundwater samples during benzene degradation and also to link the aerobic bacterial community in the groundwater sample DW3d to a specific function, i.e. benzene degradation. Sample DW3d was selected from the SIReN site for this experiment because it is heavily polluted (42.9 mgL-1; Table I) with benzene (Earle et al. 2001) and the microorganisms thriving in it have been exposed to the contaminant for a long period, suggesting adaptation to the pollutant. Furthermore, previous clone libraries indicate the presence of aerobic hydrocarbon degraders such as Polaromonas naphthalenivorans (Aburto et al. 2009).
MATERIALS AND METHODS
Study site and sampling
The study site is a BTEX–contaminated aquifer located below an operational petrochemical plant known as SIReN (Site for Innovative Research in Natural Attenuation) in the UK. This study focuses on five wells showing different physicochemical characteristics and levels of contamination, nominally classified as high, low or clean (Table I). Details about the site can be found elsewhere (Earle et al. 2001, Fahy et al. 2005).
The groundwater used to prepare the microcosms was collected on the second sampling session (19/05/05) as described previously (Aburto et al. 2009). Samples DW3s, 308s and W18s were collected with a peristaltic pump, while a bladder type was used for DW3d and W6s. Groundwater was flushed from wells until pH, temperature and conductivity were stable. Samples were collected in 1 L glass bottles. The groundwater was refrigerated and protected from light during transportation to the laboratory.
Microcosms and benzene monitoring
The microcosms were prepared approximately eight hours after sampling by dispensing 60 mL of groundwater samples DW3d, DW3s, W18s, W6s and 308s in 110 mL serum bottles, and spiking with 13C–benzene to a final concentration of 25 mgL-1, which is about half of the last measurement in situ. These groundwater samples were selected in order to compare high contaminated, low contaminated, and clean samples (Table I). Microcosms were incubated in the dark at 12 °C (which is the in situ temperature) for seven days. Autoclaved microcosms were used as controls throughout the experiment. Standards and controls all had the same liquid/headspace ratio, and were held at the same temperature as the test samples.
DNA extraction and PCR
DNA was extracted at four different time points as described previously (Aburto et al. 2009) after the preparation of the microcosms. The time points were labelled: 0, 2, 5, and 7 days that correspond to a precise sampling time of 12, 48, 120 and 168 h respectively. Partial 16S rRNA gene fragments were obtained from PCR amplification with bacterial Muyzer primers 1. Forward with GC clamp (CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG) and 2 Reverse (ATT ACC GCG GCT GCT GG) (E. coli position 341 to 534) (Muyzer 1993). The cycling conditions were as follows: an initial denaturation step of 94 °C for 1 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, and extension at 72 °C for 2 min. Cycling was completed by a final elongation at 72 °C for 10 min.
DGGE
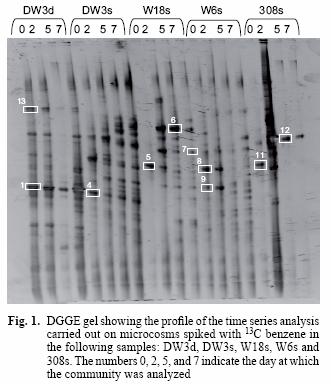
PCR products with a GC clamp were separated without purification in an 8 % w/v polyacrylamide gel with a linear denaturing gradient, increasing from 40 % at the top of the gel to 60 % at the bottom (100 % denaturants correspond to 7 M urea and 40 % v/v formamide). The DGGE gel was run at 60 °C for 5 h in a D Code Universal Mutation Detection System (Bio–Rad Hercules, CA, USA). Gels were stained for 15 min with 15 µL of SYBR Gold (Molecular Probes BV, Leiden, Netherlands) diluted in 150 mL of 1X TAE buffer. Stained gels were visualized in a GelDoc System (Bio Rad). Dominant bands were excised and incubated in 40 µL of elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA pH 8 and 0.1 % w/v SDS) for 4 h at 37 °C. DNA was precipitated with two volumes of absolute ethanol, washed with 70 % w/v ethanol and air dried. The pellet was resuspended in 20 µL DEPC treated water. Bands 1, 4, 9, 12, 13 (Fig. 1) were cloned in order to avoid sequencing co–migrating DGGE bands. The 16S rRNA gene fragment was reamplified with primer forward M13 and reverse M13 in a first instance to confirm presence of the insert and later with both forward and reverse Muyzer primers prior to sequencing and identification.
Sequencing
Sequencing reactions contained 1 µL of 10 pmol µL-1 of the reverse primer 1389R (Invitrogen), 2 µL of Big Dye Terminator V2.0 Cycle Sequencing kit, 6 µL of 2.5x sequencing buffer, 5 µL of purified PCR product (approx 15 ng) and 6 µL of water. Amplification conditions were: 25 cycles of (96 °C, 15 sec; 60 °C, 15 sec; 60 °C, 4 min). After sodium acetate and ethanol precipitation, the sequencing reaction products were run on a Perkin Elmer ABI PRISM 310 capillary electrophoresis automated genetic analyzer. Sequences were then processed with DNA Sequencing Analysis Software version 3.3.
Stable isotope probing
The protocol used in this study is an adaptation of the method used by Manefield et al. (2002a). Microcosms containing 100 mL of groundwater (prepared eight hours after sampling) from well DW3d were spiked with the stable isotope 13C–benzene (all carbon atoms labelled) to 25 mgL-1 as final concentration and incubated at 12 °C for 10 days. Nucleic acid extractions were performed at time 0 (4 hours after the 13C benzene spike) and on the days 2, 5 and 7. RNA was isolated and quantified. 50 µL of template RNA (approx. 10 ng) was added to a mixture of 9.69 mL caesium trifluoroacetate (2.0 g mL-1), 0.39 mL formamide and 1.37 mL H2O in an 11.5 mL Sorvall ultracentrifugation tube (supplied by Kendro laboratories). The mixture was centrifuged at 35,000 rpm (119,500 x g), for 48 h in a Sorvall Discovery 90SE Ultraspeed Centrifuge, with a TST 41.14 rotor. It was later fractionated in 0.5 mL aliquots by piercing the bottom of the centrifuge tube with a needle to collect the fraction, and injecting sterile H2O on top of the tube with the help of a peristaltic pump at a rate of 10 µL per second. A total of 20 fractions were collected per gradient, and refractive indices were measured. The RNA of each fraction was precipitated with isopropanol as follows: 1 mL isopropanol was added to each fraction and kept at –20 °C for 1 hour, centrifuged at 13,000 rpm for 30 min at 4 °C; the pellet was washed with 70 % ethanol, air dried and resuspended in 25 µL of DEPC treated H2O. Each fraction was later subjected to reverse transcription and PCR amplification with Muyzer primers containing the GC–clamp. Appropriate fractions were selected and run in an acrylamide gel for community fingerprinting. Bands 1 to 7 were excised, cloned and reamplified with Muyzer primers (without the GC clamp) for sequencing. Sequences were compared with those in the EMBL database using a FastA search in order to identify the closest relatives.
For clarity, from now on, a sample spiked with 13C–benzene will be called "heavy sample", while a sample with background concentrations of benzene will be referred as "light sample". In order to confirm the separation of labelled from the non–labelled RNA in sample DW3d, heavy and light gradients obtained from the isolate Rhodococcus Q71 were tested.
The Rhodococcus isolate Q71 was grown on 12C and 13C–benzene separately. Gradients were prepared with RNA extracted from both conditions. RNA fractions were reverse transcribed, amplified with Muyzer primers and run in an agarose gel.
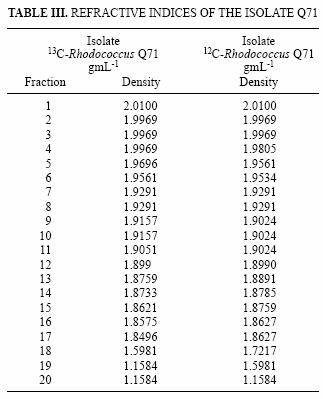
In the same way as with the Rhodococcus isolate, microcosms prepared with sample DW3d were amended separately with 12C and 13C–benzene. The gradients were centrifuged, fractionated and the refractive indices measured for each fraction (Table III). The fractions were treated as described for the Rhodococcus strain.
RESULTS
Changes in bacterial community during benzene degradation
Benzene degradation was detected from the second measurement at day 2 for most of the samples and until the final measurement on day 7 (Fig. 2) when the last microcosm was sacrificed for nucleic acid extraction. Generally, the rates of benzene degradation were similar for all samples (except the killed control), with a 60–70 % reduction in benzene concentration remaining after the 7 d incubation.
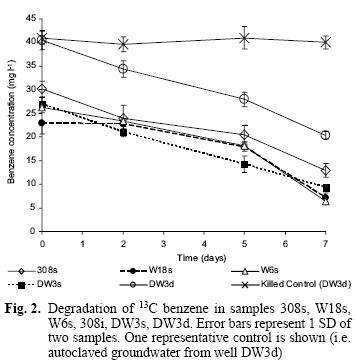
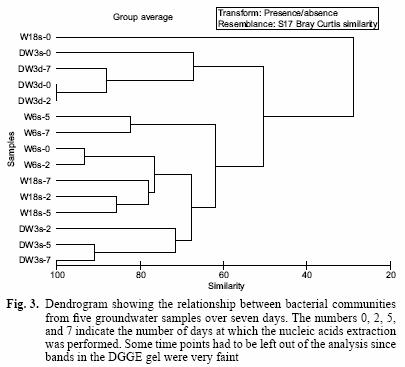
The DGGE pattern (Fig. 1) obtained from the different samples corresponds to each of the time points of benzene monitoring by gas chromatography (Fig. 2) and shows a stable community over time in most of the samples as well as different communities among the samples, which can also be seen in a dendrogram (Fig 3). All of the prominent bands in the gel were excised in order to identify them; however, several bands (n = 7) had poor sequence quality. Good quality sequences were retrieved from the bands marked on the DGGE gel (Fig. 1) and their closest relatives are shown in table II.
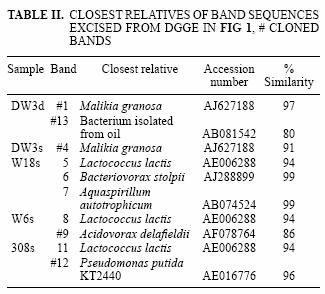
The sequences retrieved from sample DW3d were closely related (97 % similar) to Malikia granosa and a bacterium isolated from soil (Table II). The organisms retrieved from the other groundwater samples (Fig. 1) included the following bacterial species: Aquaspirillum autotrophicum (band 7) in sample W18s, Lactococcus lactis (bands 5, 8, 11) in samples W18s, W6s and 308s, Bacteriovorax stolpii (previously known as Bdellovibrio stolpii; Baer et al. 2000) (band 6) in sample W18s, and Pseudomonas putida strain KT 2440 (band 12) in sample 308s. The sequences retrieved from bands 9 (sample W6s) and 13 (sample DW3d) matched with the organisms Acidovorax delafieldii and a bacterium found in a TEX mixture respectively; however, they were distant to this organism with 16S rRNA sequence similarity below 90 % for both sequences (Table II).
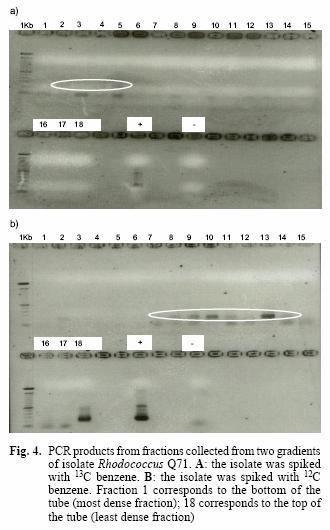
The use of stable isotope probing for detection of RNA from isolate Q71, Rhodococcus erythropolis
Stable isotope probing relies on the separation of labelled and non–labelled nucleic acids; therefore, the ultracentrifugation is critical and may be the most important step in the process. Gradients were centrifuged, fractionated and the refractive indices (Table III) indicated a good separation of the fractions.
The presence of amplicons in the first fractions (1 to 5) of the heavy sample for the isolate Q71 and their absence in the light sample suggests that the ultracentrifugation achieved the goal of separating the labelled from the non–labelled nucleic acids (Fig. 4A, B). This is supported by the appearance of bands only from the seventh fraction onwards in the light sample, except for a faint band on the second fraction. However, this band may be the product of experimental contamination as has been described previously (Manefield et al. 2002a), since no bands are visible in the adjacent fractions (1, 3, 4, 5, 6 in Fig. 4B).
The use of stable isotope probing for detection of RNA from environmental sample DW3d
On the basis of the successful results on the isolate Q71, it was decided to test the technique SIP on the environmental sample DW3d. The sample, designated as heavy, in fact contains a mixture of 13C–labelled benzene (25 mgL-1) and unlabelled benzene present in the groundwater, which is consistent with the presence of amplicons from all the fractions of the gradient. This is further supported by the lack of amplicons in the first fractions of the light sample and their presence from the sixth sample onwards (Fig 5B). Tentative evidence to support the separation of labelled and non–labelled DNA comes from the faint band in fraction 7, which is the zone where heavy and light nucleic acids are separated.
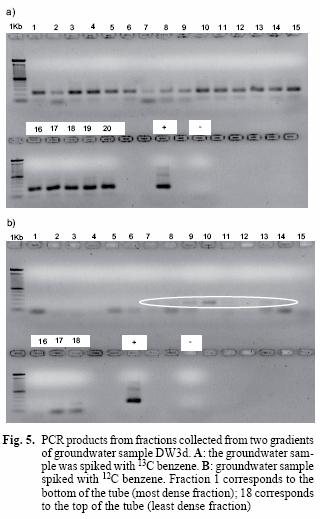
The presence of amplicons in all the fractions of the heavy sample was evident (Fig. 5A). This was expected since this sample contains heavy and light benzene; in contrast, amplicons are only visible from the sixth fraction onwards in the light sample (Fig. 5B). This suggests that fractions 1 to 5 in the heavy sample correspond to labelled DNA and is consistent with the result observed with the isolate Q71.
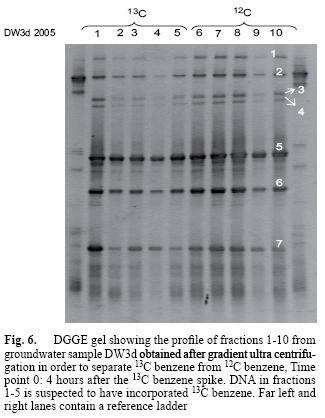
Fractions 1 to 5 and 6 to 10 of the heavy DW3d sample were assumed to represent the heavy and light DNA respectively, and were subjected to DGGE analysis. The profile obtained for both sets of fractions was very similar, and bands 1 to 7 were identical (Fig. 6). Based on 16S rRNA sequences retrieved from the bands excised from the gel, species like Malikia granosa and Acidovorax sp. KSP1 (Table IV), with sequence similarities of 92 and 91 %, respectively, were present.

DISCUSSION
Changes in bacterial community during benzene degradation
Rapid aerobic benzene degradation was observed for most of the groundwater samples (Fig. 2), which confirms the presence of adapted aerobic benzene degraders in each of the samples, in spite of the low amounts of oxygen in situ in wells DW3d, DW3s, high pH (10.2) in 308s and low benzene concentrations in samples such as W18s and W6s (Table I; Earle et al. 2001).
The DGGE profile (Fig. 1) suggests that the communities remained quite stable over 7 days, yet each groundwater community was different from each other. The relationship of these communities can also be observed in a dendrogram depicting the dissimilarity among these groundwater samples (Fig. 3). Most of the different time points cluster according to the groundwater sample and not the time point, confirming that each groundwater sample harbours different communities and that they do not change significantly over time. The only time point that was out of any cluster was W18s–0. However, this is understandable because the DGGE profile for this time point only shows one band along the lane (Fig. 1, band 5) while the successive time points for that sample include several bands in each lane. Furthermore, the rate of benzene degradation was the slowest for this sample. This suggests that the community changed dramatically in this sample from time 0 to time 2. However, it remained stable for successive time points. Samples DW3d and DW3s share five bands; one of those bands was identified as the microorganism Malikia granosa in both samples (Fig. 1, Bands 1 and 4).
Malikia granosa and Malikia spinosa are polyhydroxyalkanoate and polyphosphate accumulating bacteria (Spring et al. 2005) and are related to the Hydrogenophaga species; this is supported by the previous detection of similar clones in this groundwater sample (DW3d) (Aburto et al. 2009). Furthermore, isolates of a benzene–degrading Hydrogenophaga strain in another location (308s) of the same site have been obtained (Fahy et al. 2008). And after sequences analysis, 99 % percent similarity was shown between bands 1, 4 and the Hydrogenophaga isolate. One of the sequenced bands from groundwater sample W18s was related, with a 16S rRNA gene sequence similarity of 99 % to the organism Aquaspirillum autotrophicum. This is an aerobic hydrogen–oxidizing bacterium that contains one or more hydrogenase enzymes that bind hydrogen and use it either to produce ATP or for reducing power for autotrophic growth (Madigan 2000). This bacterium has been previously isolated from a eutrophic freshwater lake, and it can grow autotrophically or heterotrophically (Aragno 1999).
A constant band appearing at time zero for samples W18s, W6s and 308s (bands 5, 8 and 11; Fig. 1) was related, with a 94 % 16S rRNA similarity, to Lactococcus lactis. This organism, as indicated by its name, produces lactic acid as a sole fermentation product; it is an aerotolerant anaerobe able to grow even in the presence of oxygen (available in sample W18s; Table I). Another constant band appearing throughout the second to the seventh day in sample W18s was identified with a 16S rRNA gene sequence similarity of 99 % to Bacteriovorax stolpii (band 6; Fig. 1), previously known as Bdellovibrio stolpii (Baer et al. 2000). This organism is known for its ability to invade other bacteria and live parasitically on the host, until the host's death when a new generation of the B. stolpii is released (Kadouri and O'Toole 2005, Lambert et al. 2006, Rogosky et al. 2006). They can be found in diverse environments such as marine and freshwaters, sewage and soil.
A good quality sequence was obtained from sample 308s (Fig. 1; band 12); this band was closely related to Pseudomonas putida strain KT2440, which is a well known hydrocarbon degrader. This organism contains a large number of dioxygenases and at least four pathways for the degradation of hydrocarbons (Jiménez et al. 2002). Therefore, this bacterium is likely to carry out benzene degradation in spite of the high pH (10.2) and this is also supported by the isolation of benzene–degrading Pseudomonas species from the same well (Fahy et al. 2008).
It can be concluded that each of the groundwater samples harbours different communities that tend to remain stable throughout the degradation of the pollutant and that rapid benzene degradation suggests the presence of acclimated aerobic degraders in the groundwater samples studied here.
SIP
The community fingerprint of sample DW3d obtained by DGGE resulted in an identical profile for heavy and light nucleic acids (Fig 6). This is explained by the availability of both types of substrates (heavy and light) in the sample and it is clear that they were making use of both. Moreover, benzene was by far the main growth substrate present in the microcosm at concentrations far higher than any other, ruling out the possibility of another substrate for the microbes to feed on (Table I). The results in figure 6 demonstrate that those organisms appearing in DGGE profiles shortly after the onset (time point 0: 4 hours after the 13C benzene spike) of benzene degradation are likely to be directly involved in benzene degradation.

The most prominent bands were related to the strains Malikia granosa and Acidovorax sp. KSP1 (Table IV). Both species had been detected in different activated sludges elsewhere; Malikia granosa is a polyhydroxyalkanoate and polyphosphate accumulating bacteria (Spring et al. 2005) closely related to the Hydrogenophaga genera and, on this basis and the fact that several Hydrogenophaga strains have been detected and isolated from this and other groundwater samples of the same site, we strongly suggest that these bands are in fact benzene–degrading Hydrogenophaga strains identified as Malikia due to the shortness of the amplicon. Acidovorax strain KSP1 is a denitrifying bacterium capable of degrading the polyhydroxyalkanoate: poly (3–hydroxybutyrate–co–3–hydroxyvalerate) (Khan et al. 2002).
CONCLUSIONS
Fast aerobic degradation of benzene confirms the presence of acclimated microbes in situ that readily utilize the oxygen when it is available to degrade benzene; in order to support this, a few strains have been isolated from the same source (308s) (Fahy et al. 2008) as well as detected in previous clone libraries (Fahy et al. 2006). The population dynamics study shows different communities for each of the samples. As for the identification of benzene degraders by SIP in one of the samples, we succeeded in the separation of the labelled RNA and identification of the prominent bands from the heavy fractions; however, it is necessary to increase the certainty in the identification of those degraders by obtaining and sequencing a larger amplicon. Nevertheless, this study shows that stable isotope probing was useful in order to link microbial identity to function in a contaminated sample.
REFERENCES
Aburto A., Fahy A., Coulon F., Lethbridge G., Timmis K.N., Ball A.S. and McGenity T.J. (2009). Mixed aerobic and anaerobic microbial communities in benzene–contaminated groundwaters J. Appl. Microbiol. 106, 317–328. [ Links ]
Aragno M.H.G.S. (1999). The mesophilic hydrocarbon–oxidizing (knallgas) bacteria. The Prokaryotes, an evolving electronic resource for the microbiological community. Springer–Verlag release 3.14, New York, USA, 1120 p. [ Links ]
Baer M., Ravel J., Chun J., Hill R. and Williams H. (2000). A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 50, 219–224. [ Links ]
Caldwell M.E. and Suflita J.M. (2000). Detection of phenol and benzoate as intermediates of anaerobic benzene biodegradation under different terminal electron–accepting conditions. Environ. Sci. Technol. 34, 1216–1220. [ Links ]
Dean B.J. (1985). Recent findings on the genetic toxicology of benzene, toluene, xylenes and phenols. Mutat. Res. 154, 153–181. [ Links ]
Dumont M.G. and Murrell J.C. (2005). Stable isotope probing – linking microbial identity to function. Nat. Rev. Microbiol. 3, 499–504. [ Links ]
Earle R., Jones D., Lethbridge G., McCarthy P. and Thomson S. (2001). P2–208/TR/2. Project SIReN: Phase 2a. Conceptual site model & groundwater model. R&D technical report. Environment Agency, Bristol, UK, 80 pp. [ Links ]
Fahy A., McGenity T.J., Timmis K.N. and Ball A.S. (2006). Heterogeneous aerobic benzene–degrading communities in oxygen–depleted groundwaters. FEMS. Microbiol. Ecol. 58, 260–270. [ Links ]
Fahy A., Lethbridge G., Earle R., Ball A.S., Timmis K.N. and McGenity T.J. (2005). Effects of long–term benzene pollution on bacterial diversity and community structure in groundwater. Environ. Microbiol. 7, 1192–1199. [ Links ]
Fahy A., Ball A.S., Lethbridge G., Timmis K.N. and McGenity T.J. (2008). Isolation of alkalitolerant benzene–degrading bacteria from a contaminated sandstone aquifer. Lett. Appl. Microbiol. 47, 60–66. [ Links ]
Jiménez J.I., Minambres B., García J.L. and Díaz E. (2002). Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ. Microbiol. 4, 824–841. [ Links ]
Kadouri D. and O'Toole G.A. (2005). Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71, 4044–4051. [ Links ]
Kasai Y., Takahata Y., Manefield M. and Watanabe K. (2006). RNA–based stable isotope probing and isolation of anaerobic benzene–degrading bacteria from gasoline–contaminated groundwater. Appl. Environ. Microbiol. 72, 3586–3592. [ Links ]
Khan S.T., Horiba Y., Yamamoto M. and Hiraishi A. (2002). Members of the family Comamonadaceae as primary poly(3–hydroxybutyrate–co–3–hydroxyvalerate)–degrading denitrifiers in activated sludge as revealed by a polyphasic approach. Appl. Environ. Microbiol. 68, 3206–3214. [ Links ]
Lambert C., Evans K.J., Till R., Hobley L., Capeness M., Rendulic S., Schuster S., Aizawa S. and Sockett E. (2006). Characterizing the flagellar filament and the role of motility in bacterial prey–penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 60, 274–286. [ Links ]
Lueders T., Manefield M. and Friedrich M.W. (2004). Enhanced sensitivity of DNA– and rRNA–based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6, 73–78. [ Links ]
Madigan M.T.M. (2000). Brock Biology of Microorganisms. Prentice Hall, Upper Saddle River, New Jersey, 992 pp. [ Links ]
Manefield M., Whiteley A.S., Griffiths R.I. and Bailey M.J. (2002a). RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68, 5367–5373. [ Links ]
Manefield M., Whiteley A.S., Ostle N., Ineson P. and Bailey M.J. (2002b). Technical considerations for RNA–based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16, 2179–2183. [ Links ]
Muyzer G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction–amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. [ Links ]
Phelps C.D., Kerkhof L.J. and Young L.Y. (1998). Molecular characterization of a sulfate–reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27, 269–279. [ Links ]
Rogosky A.M., Moak P.L. and Emmert E.A.B. (2006). Differential predation by Bdellovibrio bacteriovorus 109J. Curr. Microbiol. 52, 81–85. [ Links ]
Rooney–Varga J.N., Anderson R.T., Fraga J.L., Ringelberg D. and Lovley D.R. (1999). Microbial communities associated with anaerobic benzene degradation in a petroleum–contaminated aquifer. Appl. Environ. Microbiol. 65, 3056–3063. [ Links ]
Sikkema J., Debont J.A.M. and Poolman B. (1995). Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59, 201–222. [ Links ]
Spring S., Wagner M., Schumann P. and Kampfer P. (2005). Malikia granosa gen. nov., sp. nov., a novel polyhydroxyalkanoate– and polyphosphate–accumulating bacterium isolated from activated sludge, and reclassification of Pseudomonas spinosa as Malikia spinosa comb. nov. Int. J. Syst. Evol. Microbiol. 55, 621–629. [ Links ]
Villatoro–Monzón W.R., Mesta–Howard A.M. and Razo–Flores E. (2003). Anaerobic biodegradation of BTEX using Mn(IV) and Fe(III) as alternative electron acceptors. Water Sci. Technol. 48, 125–131. [ Links ]














