Introduction
There are nine amino acids that human beings and mono-gastric animals cannot synthesize and need to be supplied through the diet (Galili et al., 2016). While most proteins of animal origin have suitable proportions of these amino acids, plant proteins are often suboptimal for human needs; for instance, most cereal proteins are poor in lysine and tryptophan, while those of legume origin are lower in methionine and other sulfur-containing amino acids (Mariotti and Gardner, 2019). In the developing world, prolonged consumption and over-reliance on cereal proteins can potentially lead to diseases such as pellagra, a niacin deficiency disease, and kwashiorkor, a form of protein-energy malnutrition that increases susceptibility to tuberculosis and gastroenteritis (Nuss and Tanumihardjo, 2011).
A mature corn kernel contains approximately 10 % protein and several mutants alter its amino acid profile (Zhang et al., 2018). One of them is opaque-2 (o2), a natural spontaneous mutation with soft, opaque grains, discovered in the 1920s (Singleton, 1939). Biochemical effects of o2 indicate that the homozygous recessive o2 allele increases lysine (+69 %) and tryptophan in the endosperm compared to normal maize (Mertz et al., 1964). This effect doubles the biological value of maize protein, and only with half the amount of o2 maize (relative to normal maize), the same biologically usable protein can be obtained (Bressani, 1991). In spite of its nutritional superiority, o2 maize did not become popular among farmers because the o2 gene carries some negative pleiotropic effects, including grain chalkiness, thicker pericarp, larger germ size, reduced cob weight, lower grain yield and reduced kernel weight and density (Vasal, 2000). Hence, CIMMYT made intensive efforts to improve the phenotype of o2 kernels by developing hard-endosperm maize types named QPM. Today’s QPM is interchangeable with normal maize in both phenotype and yield potential while retaining its nutritional value (Vasal, 2001).
The lysine value in QPM lines varies from 2.5 to 4.0 g per 100 g of protein (Vivek et al., 2008). Though o2 is the major determinant, a few minor modifier loci may likely work in unison with o2 to moderate the effect and alter the essential amino acid content in the homozygous (o2o2) recessive background (Vasal, 2001). In a QTL mapping study by Wang and Larkins (2001), free amino acid content in o2 maize lines was analyzed and shown to vary up to 12-fold in Oh545o2 and 3-10 times in Oh51Ao2 and W64Ao2 from non-o2 wild type counterpart. One of four loci identified was coincident with the Aspartate kinase 2 (Ask2) gene (Wang et al., 2001). This locus, located on chromosome 2, has a polymorphism that causes a single amino acid substitution in the C-terminal region of the ASK2 enzyme of Oh545o2 (Wang et al., 2007). Another study using a RIL population from the cross between B73o2 and CML161, identified QTLs for traits related to amino acid content on chromosomes 7 and 8, explaining up to 39 % of the observed phenotypic variation (Gutiérrez-Rojas et al., 2010). These studies clearly established the existence of additional modifier loci that could favorably increase accumulation of amino acid levels in QPM germplasm. Of the several loci reported, the role of Ask2 is prominent and the responsible polymorphism has been identified and characterized (Wang et al., 2007).
The cloning and characterization of the O2 locus, located on chromosome 7 (Hartings et al., 1989; Schmidt et al., 1990), followed by detection of three SSR markers viz. phi057, phi112 and umc1066 (Chin et al., 1996) led to effective differentiation of the O2 and o2 alleles in elite QPM and non-QPM germplasm (Babu et al., 2005). These o2-specific SSR markers are the foundation for marker-assisted selection (MAS) in QPM breeding (Babu et al., 2005; Gupta et al., 2013; Pukalenthy et al., 2020); however, they are not enough to achieve the full effectiveness of molecular breeding for QPM genotypes. Marker umc1066 is easily visualized on agarose gels but it is commonly not polymorphic, at least in CIMMYT breeding populations; phi057 usually requires the use of polyacrylamide gels, while phi112, which is based on a deletion in the promoter region, is a dominant marker, and hence, cannot be used in the identification of heterozygotes in F2/BC generations (Babu and Prassana, 2014). These shortcomings, coupled with low throughput and time consuming of gel-based SSR analysis, result in a major hindrance to routine and large-scale adoption of marker technology in the breeding pipelines (Xu and Crouch, 2008). This requires the identification of SNP markers, which have propelled plant molecular breeding because of their ease of automation, high throughput, low cost and high read accuracy (Rasheed et al., 2017). Though the functional SNP conferring favorable response in Ask2 has been reported (Wang et al., 2007), high throughput assays are not yet available and the interaction between o2 and Ask2 loci, involving breeding germplasm, has not been documented. The objectives of this study were: 1) to evaluate the interaction between o2 and Ask2 in an F2 population derived from tropical maize germplasm and validate synergistic effect, if any, for enhanced amino acid accumulation in the endosperm, 2) to identify widely conserved SNPs within the o2 gene that can effectively discriminate the alleles in a range of elite genetic backgrounds, 3) to estimate the frequency of favorable Ask2 allele in CIMMYT germplasm and, 4) to develop medium to high throughput marker assays to facilitate the implementation of a comprehensive MAS scheme for rapid development of QPM germplasm.
Materials and methods
Interaction analysis
Nine genotypic classes were determined after genotyping 400 seeds of an F2 population resulting from a cross between CML172 (QPM) and CML323 (non-QPM). Genotyping was carried out with molecular markers for o2 and Ask2 using DNA from endosperm. DNA extraction followed a protocol based on CTAB-lauroylsarcosine described by Gao et al. (2008). To genotype o2, SSR marker phi057 was amplified by PCR reactions of 15 µL containing 1X Taq buffer, 2.5 mM MgCl2, 150 μM each dNTP, 0.3 μM of each primer (forward 5´-CTCATCAGTGCCGTCGTCCAT-3´ and reverse 5´-CAGTCGCAAGAAACCGTTGCC-3´), 1 U homemade Taq polymerase and 20 ng of DNA. The PCR program consisted of an initial denaturation of 1 min at 94 °C followed by 30 cycles of 30 s at 94 °C, 1 min at 58 °C, 1 min at 72 °C and a final extension of 5 min at 72 °C using a MJ Tetrad thermalcycler (MJ Research, St. Bruno, Quebec, Canada). Three microliters of each PCR product was migrated in 6 % acrylamide:bis-acrylamide (29:1) gels at 250 V for 2 h in 1X TBE (90 mM Tris-borate, 2 mM EDTA pH 8.0) in a MGV-116-33 system (CBS Scientific®, San Diego, California, USA). The reference of molecular weight was ϕX174/HaeIII. After electrophoresis, the gels were silver-stained according to CIMMYT (2005).
To genotype Ask2, primers flanking the functional SNP of Ask2 (Wang et al., 2007) were designed to amplify a product of 151 pb. PCR mix included 1X Taq buffer, 2.0 mM MgCl2, 166 µM each dNTP, 0.1 µM of each primer (forward 5´-CTTGGTGGTCCATGACAGTG-3´ and reverse 5´-CTAATGGCTGTGGATTGTGC-3’), 1 U homemade Taq polymerase and 50 ng of DNA in a reaction volume of 15 µL. The PCR program consisted of one cycle of 94 °C for 1 min, 58°C for 40 s, 72°C for 1 min, and a final extension at 72 °C for ٥ min performed in a MJ Tetrad thermalcycler (MJ Research, St. Bruno, Quebec, Canada). PCR products were resolved as single-stranded conformation polymorphisms (SSCP) in 16 % polyacrylamide gels applying a modified Laemmli´s protocol that consisted of preparing each layer of the gel (stacking and resolving) without SDS (Laemmli, 1970). The PCR products were diluted with 5 µL of loading buffer (10 mM NaOH, 95 % formamide, 0.05 % bromophenol blue, 0.05 % xylene cyanol), denatured during 10 min at 94 °C and quick quenching on ice. Five μL were run for 15-16 h at 150 V and 8 °C in a MGV-116-33 system (CBS Scientific®, San Diego, California, USA) using 1X TBE as running buffer. The gels were stained with 1X SYBR® Gold (Life Technologies, Carlsbad City, California, USA) for 20 min and documented by scanning in a Typhoon 8600 imager (Amersham Biosciences Ltd., Chicago, Illinois, USA).
After genotyping, each marker was scored by hand and seeds with the same genotype were bulked to form a genotypic class. Only 20 seeds per class were used to quantify 11 amino acids by high-performance liquid chromatography (HPLC). Ethanolic extracts by HPLC (consisting of a pump P680, Dionex, autosampler 465 and fluorimetric detector SFM 25, Kontron, Eching, Germany) after derivation of the primary amino group with o-phthaldialdehyde (OPA) were carried out as described by Jarrett et al. (1986). Aliquots (35 µL) of ethanolic extracts were combined with 35 µL of OPA reagent [25 mg OPA dissolved in 500 µL methanol, the addition of 4.5 mL 0.8 M borate buffer (pH 10.4) and 50 µL of mercaptopropionic acid], mixed and kept for 2.5 min at 4 °C to allow complete derivation. The extract was injected into a pre-column (Hypersil 10 µm, Phenomenex, Torrance City, California, USA) before separation on a C18 column (Hypersil ODS, 150 × 4.6 mm, 3 µm particle size, Thermo Fisher Scientific, Bonn, Germany) maintained at constant 18 °C using a thermostatic column holder (560-CIL, ERC, Riemerling, Germany) at a flow rate of 0.8 mL min-1. Eluent A was 8 mM Na2HPO4 (pH 6.8) containing 0.4 % (v/v) tetrahydrofuran. Eluent B was a mixture of 250 mL 40 mM Na2HPO4 (pH 6.8), 150 mL methanol and 105 mL acetonitrile. Two different gradients determined major and minor amino acids. For major amino acid columns were equilibrated for 10 min with 10 % A and separation conditions were as follows: 0-5 min, isocratic elution with 0 % B; 5-15 min, linear gradient from 0 % to 15 % B; 15-15.5 min, linear gradient from 15 to 100 % B; 15.5-21 min 100 % B. For minor amino acids, columns equilibrated for 10 min with 100 % A and separation conditions were as follows: 0-5 min, isocratic elution with 0 % B; 5-16 min, linear gradient from 0 to 15 % B; 16-26 min, linear gradient from 15 to 50 % B; 26-42 min, linear gradient from 50 to 60 % B; 42-46 min from 60 to 100 % B; 46-52 min 100 % B. Amino acids were identified and quantified by comparison with original standards (Sigma-Aldrich) using Chromeleon software (version 6.8, Dionex). Individual analyses of variance per amino acid and principal component analysis were applied using PROC GLM and PRINCOMP of SAS, respectively (SAS Institute, 2002).
Polymorphism identification
Fifteen elite inbred lines were chosen to search for polymorphisms within the o2 gene. Seven inbred lines were QPM, of diverse agro-ecological zones, with excellent kernel modifying capacity and of extensive use in the QPM conversion programs. The other eight lines were non-QPM, with wide adaptability and superior performance in Asia, Latin America and East African highlands (Table 1). Leaves of four seedlings per line were used separately to extract genomic DNA following a protocol based on CTAB for small-scale isolation of high quality DNA (CIMMYT, 2005). Two sets of primers were designed to amplify two genomic fragments of o2 (477 and 530 pb) and to identify regions with high degree of conservation. The reference sequence X15544, which belongs to the zein regulatory gene o2 of Zea mays (http://www.ncbi.nlm.nih.gov/), plus six cDNA sequences of o2 gene (AJ41297, AJ491298, AJ491299, AJ491300, M29411 and X16618) were used as templates for the design of primers O2F1: 5´-CTAGTGTTTGCTTCTCCCTTCC-3´, O2R1: 4´-TCCTCAGTATGGCATTGTACTCC-3´ and O2F3: 5´-TGTTGTGACCTCAGATCAACG-3´, O2R3: 5´-TTCCAGTTCTTTCAGGTGAGC-3. The PCR reactions were optimized to produce single bands without smear by applying a lower number of cycles and a large volume of reaction (30 µL). In total, two PCR fragments per seedling were amplified and purified with Qiagen spin columns (Qiagen®, Hilden, Germany), and cloned into pGEM®-T Easy Vector (Promega, USA) according to manufacturer recommendations. Recombinant plasmids were selected after transforming DH5α competent cells of E. coli by heat-shock according to CIMMYT (2005). Plasmid DNA extraction was with Wizard® Plus SV Minipreps DNA Purification System (Promega, USA) and verification of inserts was by digestion with EcoR1. Two plasmids of each genotype were sequenced at CINVESTAV facilities (Guanajuato, México), aligned using Clustal X version 1.8 (Thompson et al., 1997) and refined by hand.
Table 1 Maize inbred lines used to identify polymorphisms in the o2 locus.
| Grain color and adaptation zone | QPM line | Non-QPM line |
| White, tropical lowland | CML159 | CML343 |
| CML491 | CML495 | |
| CML502 | CL RCW22 | |
| CML492 | ||
| Yellow, tropical lowland | CML161 | CML348 |
| CML165 | ||
| White, sub-tropical | CML176 | |
| White, highland | CML244 | |
| CML349 | ||
| White, Ethiopian highland | F7287 | |
| A7018 |
Markers assays validation
A panel of 88 inbred lines from the Maize Program or CIMMYT Genbank was used to validate polymorphisms using genomic DNA as template. DNA extraction protocol was the same as that used for polymorphism identification. Two SNPs were assayed at LGC Biosearch Technologies (https://www.biosearchtech.com/) using the KASP™ chemistry. One of them was the o2-specific SNP, which differentiated QPM and normal lines and the other was the Ask2-specific SNP amplified to determine genotypic classes. This last one was also screened as SSCP marker to explore the natural variation of Ask2 and to estimate the frequency of the favorable allele in CIMMYT germplasm.
Results
Interaction analysis between o2 and Ask2
The interaction between o2 and Ask2 was studied in a tropical germplasm background by developing a segregating population from a cross between CML172, which is a QPM line possessing favorable alleles for both o2 and Ask2 loci (based on ‘2’ type SSCP conformer), and CML323, a non-QPM line. The interaction analysis was based on the concentration of 11 amino acids of nine genotypic classes (Table 2). Among these classes, the double recessive homozygous class (G1) contained significantly higher amounts of lysine, tryptophan and aspartic acid relative to the double dominant homozygous class (G5). In this regard, G1 had 2.3-fold more lysine, 1.3-fold more tryptophan and 5.3-fold more aspartic acid than G5. Lysine levels of G1 were also 60 % higher relative to G2, the recessive dominant class. A fast analysis using biplot analysis indicated that the first two components explained 96.24 % of the variance (Figure 1). PC1 explained 89.27 % of variation while PC2 explained 6.93 %. All vectors had a positive direction toward G1, G2 and G3 where o2 was recessive. No vector pointed toward genotypes with the dominant allele of o2 (G4 to G9). On the contrary, Ask2 locus on its own was not found to have any significant effect on any of the amino acids analyzed, but appeared to interact synergistically with o2 and it significantly enhanced lysine, histidine and methionine levels in the double recessive homozygous class (G1).
Table 2 Amino acids concentration (nmol g-1 FW) of kernels of the genotypic classes of o2 and Ask2 in CML172 × CML323.
| Genotypic class | o2 | Ask2 | Amino acids | ||||||||||
| Asp | Glu | Asn | Thr | His | Met | Trp | Phe | Ile | Leu | Lys | |||
| CML172 | 1 | 1 | 4969.74 | 2759.53 | 4631.05 | 286.31 | 61.10 | 27.98 | 53.14 | 210.12 | 159.79 | 268.78 | 719.06 |
| CML323 | 2 | 2 | 268.04 | 329.00 | 656.81 | 64.00 | 41.36 | 39.11 | 45.95 | 36.17 | 41.80 | 55.04 | 81.75 |
|
|
|||||||||||||
| G1 | 1 | 1 | 3383.40 | 1203.85 | 1469.11 | 297.81 | 230.78 | 100.67 | 105.13 | 155.70 | 199.92 | 224.46 | 431.92 |
| G2 | 1 | 2 | 3095.29 | 1294.23 | 2312.75 | 349.47 | 160.47 | 47.47 | 112.76 | 156.63 | 192.91 | 249.35 | 270.02 |
| G3 | 1 | H | 1979.50 | 1108.96 | 1189.26 | 306.18 | 120.90 | 60.67 | 66.57 | 112.90 | 169.72 | 250.83 | 364.71 |
| G4 | 2 | 1 | 409.54 | 393.07 | 351.05 | 179.56 | 68.63 | 31.10 | 37.67 | 58.01 | 81.12 | 171.76 | 94.02 |
| G5 | 2 | 2 | 541.97 | 557.88 | 589.25 | 222.23 | 98.03 | 36.49 | 46.35 | 67.43 | 94.24 | 177.15 | 132.78 |
| G6 | 2 | H | 460.78 | 366.26 | 374.36 | 180.02 | 104.14 | 38.35 | 49.94 | 48.73 | 62.50 | 129.35 | 124.24 |
| G7 | H | 1 | 298.44 | 247.67 | 309.08 | 149.93 | 73.33 | 32.13 | 37.31 | 46.06 | 46.36 | 103.12 | 102.82 |
| G8 | H | 2 | 471.34 | 342.96 | 385.82 | 170.18 | 95.32 | 39.26 | 38.24 | 50.65 | 72.13 | 128.60 | 126.01 |
| G9 | H | H | 315.77 | 300.76 | 300.61 | 159.87 | 88.22 | 31.29 | 39.22 | 43.81 | 57.83 | 114.87 | 96.99 |
| o2 classes | |||||||||||||
| G1,G2,G3 | 1 | - | 2819.40a | 1202.35 a | 1657.04 a | 317.82 a | 170.72 | 69.61 | 94.82 a | 141.74 a | 187.52 a | 241.55 a | 355.55 a |
| G4,G5,G6 | 2 | - | 470.76b | 439.07 b | 438.22 b | 193.94 b | 90.27 | 35.31 | 44.66 b | 58.05 b | 79.29 b | 159.42 b | 117.01 b |
| G7,G8,G9 | H | - | 361.85b | 297.13 b | 331.83 b | 159.99 c | 85.62 | 34.23 | 38.25 b | 46.84 b | 58.77 b | 115.53 b | 108.61 b |
| Ask-2 classes | |||||||||||||
| G1,G4,G7 | - | 1 | 1363.79 | 614.86 | 709.74 | 209.10 a | 124.25 | 54.64 | 60.04 | 86.59 | 109.13 | 166.45 | 209.59 |
| G2,G5,G8 | - | 2 | 1369.53 | 731.69 | 1095.94 | 247.30b | 117.94 | 41.08 | 65.78 | 91.57 | 119.76 | 185.04 | 176.27 |
| G3,G6,G9 | - | H | 918.69 | 591.99 | 621.41 | 215.36b | 104.42 | 43.44 | 51.91 | 68.48 | 96.68 | 165.02 | 195.31 |
Genotype nomenclature: 1: double homozygous recessive, 2: double homozygous dominant, H: heterozygous. Asp: aspartic acid, Glu: glutamic acid, Asn: asparagine, Thr: threonine, His: histidine, Met: methionine, Trp: tryptophan, Phe: phenylalanine, Ile: Isoleucine, Leu: leucine, Lys: lysine. Means with different letters within columns are statistically different (Tukey, P ≤ 0.05).
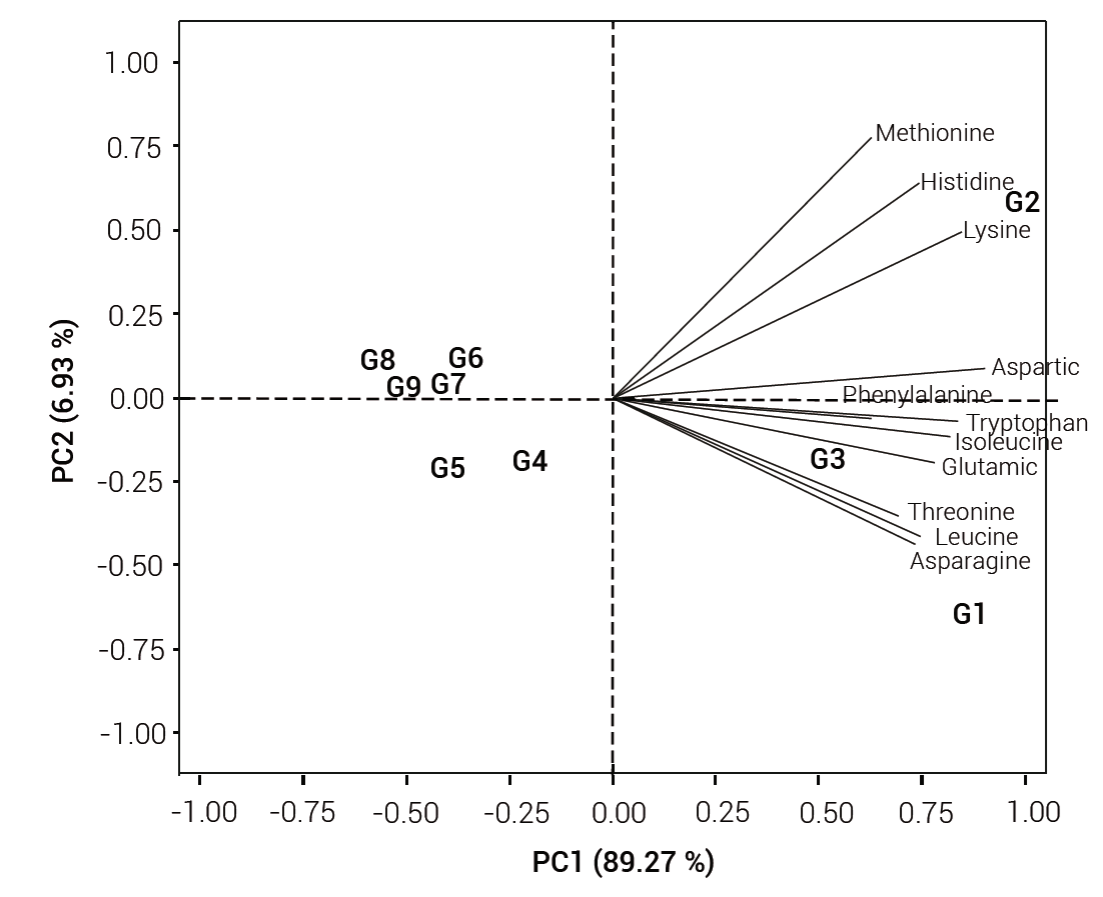
Figure 1 Biplot of principal components of 11 free amino acids content in nine genotypic classes (G) from an F2 population between CML172 (QPM) and CML323 (non-QPM). G1: o2/o2|ask2/ask2, G2: o2/o2|Ask2/Ask2, G3: o2/o2/Ask2/ask2, G4: O2/O2/ask2/ask2, G5: O2/O2/Ask2/Ask2, G6: O2/O2/Ask2/ask2, G7: O2/o2/ask2/ask2, G8: O2/o2/Ask2/Ask2, G9: O2/o2/Ask2/ask2.
Polymorphism identification in the o2 locus
Sequencing the amplicons of o2 allowed to locate an important discriminatory SNP in the region of exon 1 (Figure 2). The sequencing included part of the 5’ flanking region, four exons (1, 2, 3 and 4) and two introns (2 and 3). In total, a length of 1 kb of the genomic sequence was analyzed, 65 polymorphic sites were identified, and one polymorphism each 30 bp was averaged. In general, all QPM lines had the same sequence, indicating a possible common donor of o2 in CIMMYT lines.
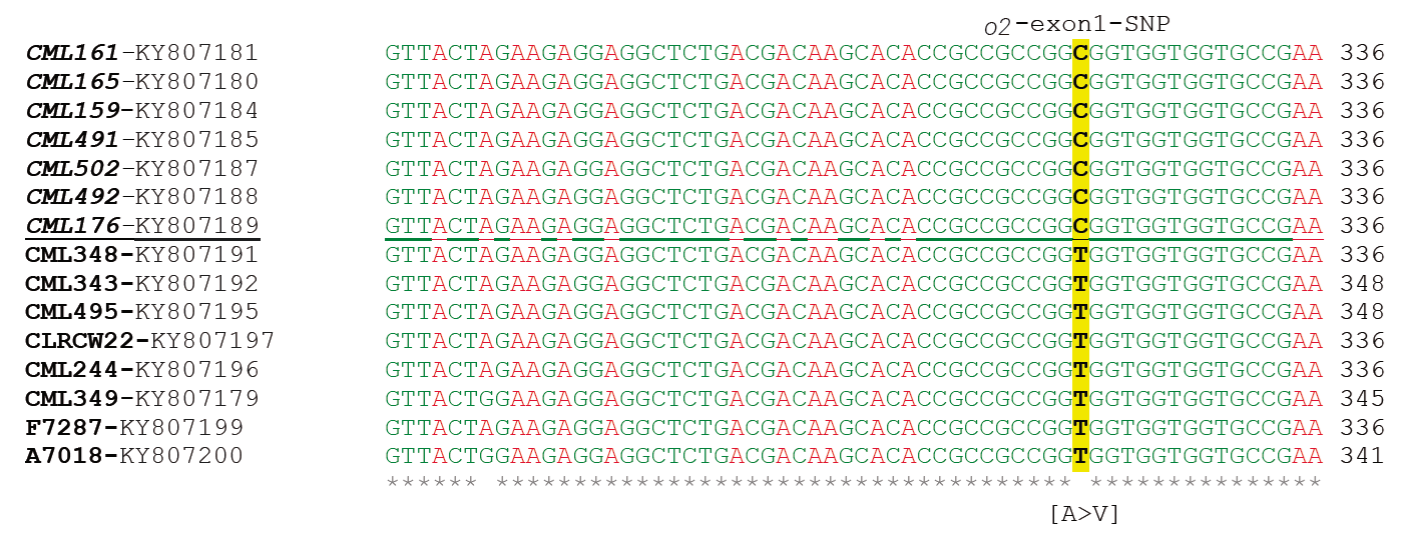
Figure 2 The o2-exon1-SNP and its context sequence. The SNP is highlighted in yellow. Allele C discriminates to QPM lines (in italics). Allele T discriminates non-QPM lines (no italics). Name of each maize line plus its GenBank ID is to the left of the nucleotide sequence.
Of the several polymorphisms that differentiated the QPM lines from non-QPM, the following four merit attention: 1) two SNP in intron 2 (positions 10,800,027 and 10,800,032 bp concerning to o2 reference sequence of Zea mays, ID 542375 of B73 RefSeq_v3) which defined three alleles with the haplotypes GT/AG/GT (donor/recipient/recipient), 2) two consecutive SNP in intron 2 (positions 10,800,038 and 10,800,039) which defined the alleles AT/TC/AC (donor/recipient/recipient), 3) two InDels in intron 2 (positions 10,799,978 to 10,799,982 and 10,799,988 to 10,799,990), which corresponded to 5-base and 3-base deletions in all QPM donor lines as well as for two non-QPM lines, and 4) an SNP of transition type in exon 1 (position 10,799,047 according to sequence ID 542375 of B73 RefSeq_v3, and position 1790 according to accession X15544 of the GenBank). The SNP corresponds to C in QPM lines and to T in non-QPM lines. This SNP was named 2-exon1-SNP, it causes an amino acid substitution (valine to alanine) and was selected for the subsequent high throughput assays. Sequences with this SNP were submitted to the GenBank of NCBI under accessions KY807178 to KY807200.
Validation of high throughput marker assays
Two high throughput KASP™ assays non gel-based were developed to facilitate large-scale MAS in pedigree/back-cross breeding programs involving QPM × non-QPM lines. One of these assays, designed to screen the o2-exon1-SNP, was firstly tested in 16 lines. The analysis of allele calls clearly grouped all genotypes into two clusters -one corresponding to allele C of QPM and another corresponding to allele T of non-QPM lines. Thus, this analysis was extended to a larger set of 88 lines in which 60 lines were QPM and the remaining non-QPM. Allele calls for o2-exon1-SNP differentiated two clusters again (Figure 3A), the cluster in red color corresponding to lines with the allele C and the cluster in blue color to lines with the allele T. The cluster with allele C included 53 QPM lines while the cluster with allele T comprised 35 lines, of which 28 were normal and seven were QPM. CML145, CML167, CML168, CML177, CML180, CML184 and CML188 were the QPM lines that possessed the ‘normal’ allele (T), contrary to expectation. Therefore, prediction value of o2-exon1-SNP was 92 % for the QPM trait.
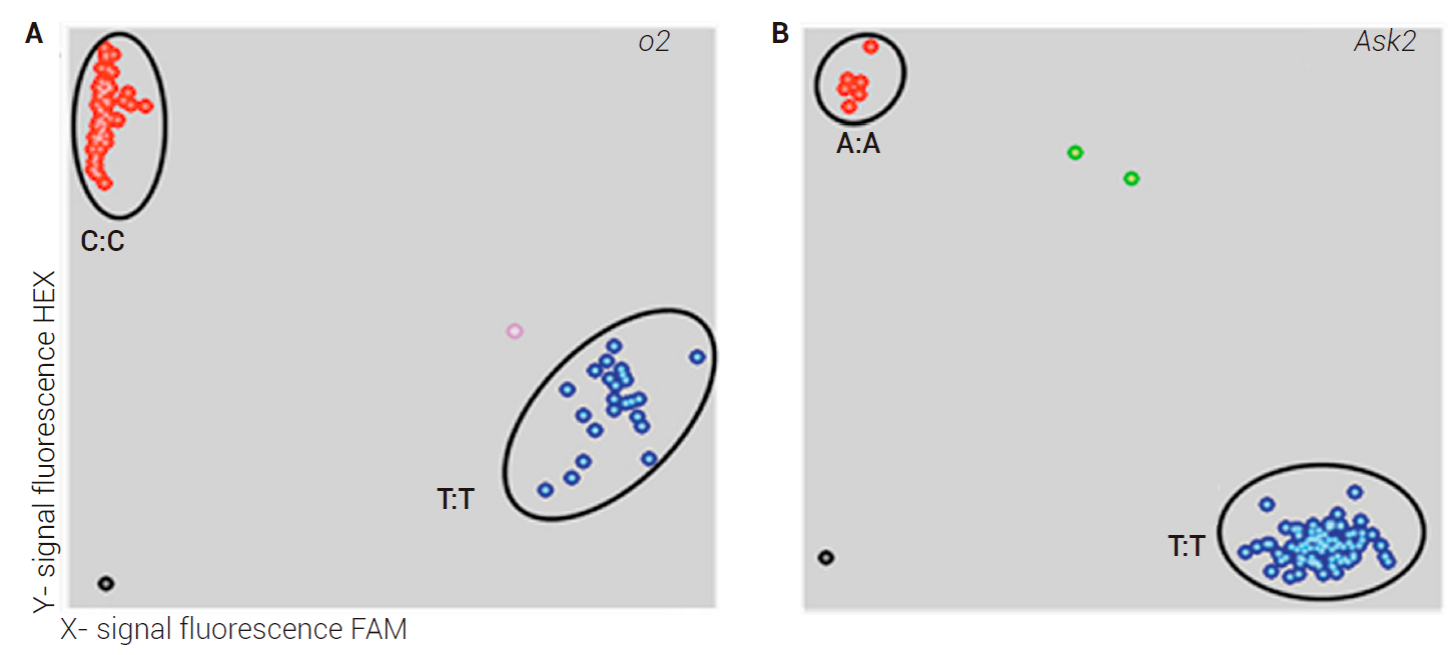
Figure 3 Genotyping cluster plot of KASP™ assays for o2 (A, left) and Ask2 (B, right) SNP. Each data point represents an individual inbreed line. Similar fluorescence values mean the same genotype. Red dots: homozygous for allele labeled with HEX, blue dots: homozygous for allele labeled with FAM, green dots: heterozygous, black dots: no template control.
The remaining assay was designed to test the functional polymorphism A (‘lysine enhancing’) of the C-terminal region of the ASK2 protein of the Oh545o2 maize line (Wang et al., 2007), which identified several lines possessing the favorable allele (Figure 3B). In this case, allele calls distinguished one cluster (in red) of seven inbred lines with the favorable allele A, of which five were QPM (CML148, CML161, CML162, CML179 and CML188) and two normal (CML328 and CML 370). The other cluster with unfavorable allele T (in blue) comprised 81 lines of which 55 were QPM and 26 non-QPM, indicating a low frequency of this allele in CIMMYT germplasm.
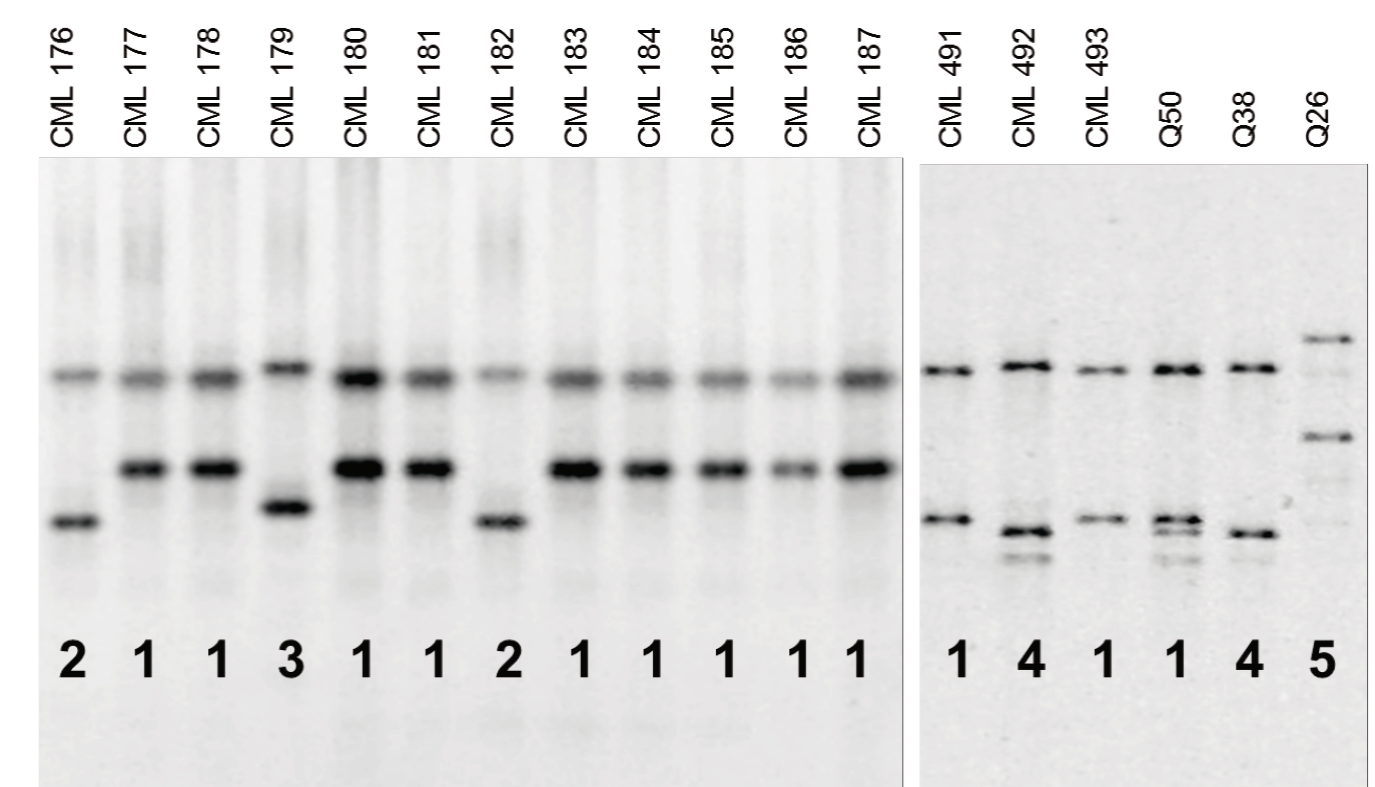
Natural variation of Ask2
Given that the favorable allele A at the Ask2 locus was only detected in five out of 60 QPM lines, to determine other possible functional variants of Ask2, a gel-based SSCP assay was developed and tested in 82 maize lines. Five distinct SSCP band patterns were unambiguously identified (Figure 4). SSCP variants were as follows: variant 1 in 60 lines (46 QPM and 14 normal), variant 2 in 11 lines (10 QPM and 1 normal), variant 3 in 5 lines (all QPM), variant 4 in 5 lines (all normal) and variant 5 in 1 line (QPM), suggesting that other polymorphisms could be involved in the high lysine level in the endosperm.
Discussion
The interaction between o2 and Ask2 in tropical germplasm background was studied in nine different genotypic classes resulting from an F2 segregating population. The single locus tests established the overriding effect of o2 on most of the amino acids, irrespective of the genotypic constitution at Ask2 locus (Figure 1). On the contrary, Ask2 locus on its own was not found to have any significant effect on any of the amino acids analyzed but did interact synergistically with o2 and significantly enhanced lysine, histidine and methionine levels in the double recessive homozygous class (G1).
The elevated levels of lysine in o2 endosperm are due to enhanced synthesis of lysine-containing proteins (Zhang et al., 2018) as well as high free lysine content (Wang and Larkins, 2001). Lysine is synthesized through a branch of aspartate pathway and is regulated by feedback inhibition loops, in which the accumulating lysine suppress the activity of two key enzymes, aspartate kinase (AK) and dihydrodipicolinate synthase (DHDPS) (Galili, 1995; Galili et al., 2016). The native mono-functional AK enzyme is a tetramer composed of two α- and two β-subunits, encoded by Ask1 and Ask2 genes, respectively (Dotson et al., 1989; 1990). Mutant alleles in either Ask1 or Ask2 loci result in AK enzyme that is less sensitive to feedback inhibition and lead to overproduction of lysine, threonine, methionine and isoleucine (Dotson et al., 1990). In maize, Ask2 explains a significant portion of variance for the free lysine levels in the o2 endosperm (Wang and Larkins, 2001), and thus, it represents an important target for enhancing free lysine levels in seeds. In this study, the mutation in the Ask2 locus that resulted in a favorable SSCP allele in CML172 is either insensitive or less sensitive to lysine inhibition and, thereby, contributed to increased accumulation of lysine and other aspartate pathway amino acids, similar to the mutant Ask2 allele of Oh545o2; however, it was apparent that the Ask2 locus on its own did not have any significant impact on the lysine or other amino acid levels in this investigation. Based on studies involving double mutants of Ask1 and o2, Brennecke et al. (1996) inferred that Ask1 is being directly or indirectly regulated by O2 because AK in these double mutants is less sensitive to lysine feedback inhibition than that of Ask1 mutation alone. Though there is no direct evidence for Ask2 being regulated by O2, such a possibility may partially explain the insignificant effect of Ask2 locus independent of o2.
Though the three o2-specific SSR markers offer an excellent opportunity for MAS, their use has a limited potential in plant breeding. In this investigation, an important discriminatory SNP in the exon 1 region of o2 was identified and an easy to use, high throughput assay, non-gel-based to facilitate large-scale MAS was developed. The lack of a perfect correlation between the underlying allelic state at this SNP locus and the QPM status indicates the non-causal nature of this variation, which however has a high predictive value for the QPM trait. Assays SSCP and KASP were designed for Ask2 which are able to screen a large collection of QPM and normal lines in the CIMMYT germplasm and to identify several lines possessing the favorable allele. Interestingly, the frequency of the unfavorable allele at the Ask2 locus was significantly higher in the QPM germplasm compared to normal endosperm, which could be due to the high selection pressure exerted by breeders during the process of QPM germplasm development through rigorous biochemical tests for high lysine and tryptophan. The higher free amino acid content in the o2 endosperm could either be due to increased lysine synthesis or loss of lysine ketoglutarate reductase (LKR) activity, which degrades lysine as endosperm matures (Arruda et al., 2000). Apart from Ask2, several genes have been postulated to play key roles in the enhanced lysine synthesis such as aspartate kinase-homoserine dehydrogenase-2 and dihydro dipicolinate synthase (Galili et al., 2016; Wang and Larkins, 2001). Some of such loci may likely be at play in those QPM lines that are rich in lysine but do not have the favorable Ask2 allele. Identifying and characterizing such loci in the high lysine QPM lines may provide further incremental gains on the enhanced amino acid levels conferred by o2 and Ask2.
The gel-based SNP detection assay developed in this investigation for the Ask2 locus combines reasonable throughput of modern gel electrophoresis platforms with robust discriminative power of the SSCP technique (Orita et al., 1989). The SSCP-SNP assay is efficient and sensitive enough for most of the PCR products in the size range of 100-750 bp, cost-competitive and requires no sophisticated instrumentation (Sunnucks et al., 2000). Notably, some of the lines (e.g. CML172) that were identified as possessing favorable allele at the Ask2 locus (‘2’ type), as per SSCP conformers, had indeed the unfavorable allele (T) according to KASP genotype calls. This discrepancy between SSCP and KASP genotypes in the Ask2 locus was possibly due to other unknown base changes in the Ask2 amplified region. Based on the results of these genetic analyses in the segregating F2 progenies, it was demonstrated that unknown base changes in the exon region of Ask2 may also confer positive responses (lysine enhancing) similar to the previously established functional SNP. It is indeed, possible to obtain higher genetic gains for enhanced lysine and tryptophan levels if the QPM lines identified in this investigation, that harbor favorable alleles (either A as per KASP assay or ‘2’ type SSCP conformer) at the Ask2 locus, are preferred as QPM donors in the new line conversions or pedigree starts and tracked through MAS in the subsequent multiple filial generations.
With the development of an array of low-cost and high-throughput genotyping platforms (Semagn et al., 2014), MAS has emerged as an attractive proposition in most commercial and large-scale public-sector maize breeding programs and quickly becoming a compelling option especially ever since doubled haploids are integrated as a mainstream breeding tool in the maize improvement pipeline (Chaikam et al., 2019; Maqbool et al., 2020). Four possible ways are envisioned in which the high throughput SNP markers developed in this investigation for o2 and Ask2 could be deployed in the QPM breeding programs: 1) as an inexpensive allele mining tool to identify putative donor lines in large and diverse germplasm collections, including non-adapted genotypes and wild relatives that have never been subjected to biochemical tests, 2) as an informed choice of parental lines based on the favorable allele composition and, consequently, as a predictive tool to cut down the number of crosses in QPM breeding program, 3) to fix the favorable alleles at o2 and Ask2 loci in homozygous condition at an early stage of recombination such as F2/BC1/BC1F2 in pedigree/backcross breeding schemes, and 4) as fixed effect covariates in a genomic selection index to efficiently predict the likely high-lysine/tryptophan entries among the biochemically untested germplasm such as conventionally bred material or DH lines that are generated routinely in large numbers in the breeding program.
Conclusions
Results pointed to an independent effect of o2 and Ask2 rather than an interaction effect between both loci concerning to amino acids content in the maize endosperm. The o2 variant alone provided a highly significant effect for high accumulation of amino acids in the double recessive genotypes and this accumulation could be positively influenced and modified for lysine, histidine and methionine by Ask2. Evidence is provided from high-throughput assays that could enhance the efficacy of marker selection for QPM germplasm development. One of them, the SNP of exon 1 of the o2 locus had a highly predictive diagnostic value for the QPM trait, while the other one, the SSCP marker led to the identification of genetic variants in QPM germplasm that opens the opportunity to find other polymorphisms associated with the high accumulation of lysine.











 nueva página del texto (beta)
nueva página del texto (beta)