Introduction
The economic impact of bacterial diseases in crops can be large. The genus Pseudomonas comprises gram-negative plant pathogenic species, with P. syringae being the most economically important with more than 50 pathovars (Höfte and De Vos, 2006). The bacterial genus Clavibacter consists of only one species C. michiganensis which is a gram-positive phytopathogen and is subdivided into five subspecies, according to their host specificity. Both genera include important tomato pathogens: P. syringae pv. syringae, P. syringae pv. tomato and C. michiganensis subsp. michiganensis (Eichenlaub et al., 2006).
In the field, to combat bacteria there is no alternative other than the application of pesticides, so it remains as the principal available method for control of pathogens. This has significantly increased the concentration of pesticides in food and in our environment, with associated negative effects on human health (Tago et al., 2014). It is then clear that more sustainable methods are required to fight plant diseases. Recent advances in genomics and molecular genetics of host resistance and pathogenesis give hope of novel solutions to many important crop diseases (Wulff et al., 2011).
Plants possess a well-adapted innate immune system consisting of two perception systems: PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Walters, 2015). In PTI, cell surface-located receptors, called patternrecognition receptors (PRR), detect conserved pathogenassociated molecular patterns (PAMP), which act as a signature of a whole class of microbes. Pathogens have evolved mechanisms to evade PTI and deliver proteins, called effectors, directly into the cell (Boller and Felix, 2009). These effectors are mostly species-specific and promote virulence leading to effector triggered susceptibility (ETS). If the plant recognizes the effector, ETI is then activated. It mostly acts inside the cell, using the nucleotide-binding leucine-rich repeat (NB-LRR) protein products encoded by resistance (R) genes (Walters, 2015). ETI is race-specific and rarely confers broad-spectrum disease resistance. Moreover, it is often rapidly overcome by evolving pathogens that lose or mutate the nonessential recognized effector or that produce new effectors to counteract ETI (Lacombe et al., 2010). When PRRs detect PAMPs a cascade of signaling pathways is triggered, which cause activation of defense mechanisms and ultimately leads to either resistance or cell death.
One of the best-characterized PRRs is the receptor-like kinase (RLK) FLAGELLIN-SENSING 2 (FLS2). The Arabidopsis FLS2 (AtFLS2) receptor recognizes the highly conserved 22-amino-acid epitope (flg22) of the flagellin protein. For an effective activation of the PTI outputs, FLS2 has to interact with co-receptors which are members of the somatic embryogenesis-related kinase (SERK) family, mainly BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) and BAK1LIKE 1 (BKK1) (Sun et al., 2013). Another well described PRR is the RLK EF-Tu RECEPTOR (EFR), which recognizes the bacterial elongation factor EF-Tu. The receptor detects the N-acetylated peptide comprising the first 18 aminoacids of EF-Tu, termed elf18. Perception of elf18 triggers an oxidative burst, callose deposition, seedling growth inhibition and defense gene expression ultimately leading to resistance to infection with pathogenic bacteria (Kunze et al., 2004). Just like FLS2, EFR is dependent on co-receptor BAK1 to trigger PTI outputs.
BAK1 (also called SERK3), is a LRR-RLK of the SERK family. It positively regulates brassinosteroid (BR) responses, such as cell elongation and division, by forming a ligand-dependent complex with the BR receptor BRI1 (He et al., 2013). BAK1 also plays a role in plant PTI, working as co-receptor for FLS2, EFR and other PRRs (Chinchilla et al., 2009). Recent results have demonstrated the biotechnological potential of PRRs in providing enhanced immunity. Many major resistance genes have now been shown to retain function when transferred between species. The EFR pattern-recognition receptor, present only in Brassicaceae, functions to provide bacterial disease control in Solanaceae (Lacombe et al., 2010; Wulff et al., 2011; Zipfel et al., 2006).
The deployment of several genes within a single variety is known as pyramiding (Kumar and Nayak, 2010). In theory the likelihood of a pathogen evolving to overcome a gene pyramid is reduced in proportion to the number of its component resistance genes (Fukuoka et al., 2009). Based on the above, tomato plants (Solanaceae) were transformed with a cassette constituted by a pyramid of three Arabidopsis thaliana genes related to PTI (AtEFR, AtFLS2 and AtBAK1), in order to provide the plants with a broad-spectrum resistance against bacteria. It was hypothesized that plants expressing multiple PAMPs recognition receptors (PRRs) will show resistance to a wide range of bacteria, and that this resistance will be hard to overcome by pathogens, and therefore, more durable.
Materials and methods
The EFR, FLS2 and BAK1 genes were amplified by PCR from a cDNA library of A. thaliana ecotype 'Columbia' using the primers (EFR: 5'-ATCGGGTACCATGAAGCTGTCCTTTTCACTTG-3' and 5'-ATCGGCATGCCATAGTATGCATGTCCGTATTTAAC-3'; FLS2: 5'-ACTGGTCGACATGAAGTTACTCTCAAAGAC-3' and 5'-ATCGGGATCCAACTTCTCGATCCTCGTTACG-3'; BAK1: 5'-ACTGTCTAGAATGGAACGAAGATTAATGATC-3' and 5'-ATCGCTCGAGTCTTGGACCCGAGGGGTATTC-3'), where underlined bases indicate restriction sites added for gene subcloning. The AtEFR, AtBAK1 and AtFLS2 genes were linked in that order using endonuclease restriction and ligase enzymes. Genes were separated from each other by an ubiquitin monomer from tobacco. The final cassette composed by the three genes was cloned into the plant expression vector pCAMBIA 2301. This vector has the Cauliflower Mosaic Virus 35S promoter, in addition to β-glucuronidase (GUS) and the kanamycin resistance genes.
Plasmid pCAMBIA-EFR:Ub:BAK1:Ub:FLS2 containing the PRRs genes cassette was transferred into Agrobacterium tumefaciens strain-LBA4404 by electroporation. Transgenic tomato (S. lycopersicum) variety TA234 plants expressing pCAMBIA-EFR:Ub:BAK1:Ub:FLS2 was generated by the routine method used in the laboratory as described by Peña-Ramírez et al. (2007). Several primary transformants were recovered after selection on kanamycin-containing plant plates. After transfer to soil, the kanamycin resistant plants were tested for GUS activity, resulting in four positive plants, which were grown in the greenhouse and allowed them to self-pollinate to get F1 to F4 generations. GUS assays were performed on plants from each generation.
Genomic DNA was extracted from young leaves of GUS positive plants from each generation using Plant DNAzolTM reagent (InvitrogenTM). PCR was performed on those DNAs using the same set of primers employed to amplify the AtEFR, AtBAK1 and AtFLS2 genes from the A. thaliana cDNA library.
Total RNA was extracted from young leaves of each plant generation using PureLink® Plant RNA Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and cDNA was synthesized from the RNA using SuperScript® III Reverse Transcriptase (InvitrogenTM). Reverse transcription PCR (RT-PCR) was performed on the cDNA using the primers (EFR: 5'-GGCGATTATAACCTCCACAG-3' and 5'-TACTGCTTCATCCGTTCTC A-3'; FLS2: 5'-ACGCCTCTGATCTAATGGG-3' and 5'-GGATGACTCTGGTTCTCTTCG-3'; BAK1: 5'-TGACGCTACAAGTTCTGGAT-3' and 5'-ATGGCGGTGTAGGAGAGATA-3'; EF-α 5'-TACTGG TGGTTTTGAAGCTG-3' and 5'-AACTTCCTTCACGATTTCATCATA-3').
For the bacterial challenge tests S. lycopersicum variety TA234 was grown from the transgenic lines generated as one plant per pot at 22 oC under a 16 h photoperiod. Pseudomonas syringae pv. syringae B728a and Pseudomonas syringae pv. tomato DC3000 were grown in a liquid mannitol glutamate (MG) medium (10 g L-1 mannitol, 2 g L-1 L-glutamic acid, 0.5 g L-1 KH2PO4, 0.2 g L-1 NaCl, 0.2 g L-1 MgSO4·7H2O, pH 7.0) at 28 oC. Bacterial suspensions were prepared as previously described by Katagiri et al. (2002), and were adjusted to 1 x 105 colony-forming units (CFU) mL-1 (OD600 = 0.0002) in sterile phosphate (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4).
Three-week-old tomato plants with three to four true leaves were used for inoculation. Bacterial suspensions were infiltrated with a needleless hypodermic syringe into the leaves. As a control, plants were mock inoculated in the same way with PBS. Bacterial growth was monitored within leaf tissue by grinding six leaf disks (0.6 cm2) per sample and plating dilutions of the ground material on King's B (KB) medium (20 g L-1 peptone, 1.5 g L-1 MgSO4·7H2O, 1.5 g L-1 K2HPO4, 15 mL L-1 glycerin, pH 7.2) with 25 μg mL-1 nalidixic acid at 28 oC. Three replicated samples were taken every other day over a 10 d period. Each experiment was independently conducted three times within each plant generation.
For Clavibacter michiganensis subsp. michiganensis, bacterial cultures were prepared and inoculations were performed as described by Louws et al. (1998). Briefly, Cmm was grown in liquid nutrient broth yeast extract (NBY) medium (8 g L-1 nutrient broth, 2 g L-1 yeast extract, 2 g L-1 K2HPO4, 0.5 g L-1 KH2PO4, 5 g L-1 glucose, pH 7.0) at 28 oC. Bacteria were suspended into sterile PBS buffer and adjusted to 2 x 108 CFU mL-1 (OD600 = 0.16). Three week old tomato plants with three to four true leaves were used for inoculation. For inoculation, a scalpel dipped in inoculum was used to make a tiny wound on the stem of the plant, between the cotyledons. As a control, plants were mock inoculated in the same way with sterile PBS.
Every 5 d, from day 20 to day 40, a 1 cm segment of the stem 10 cm above the inoculation site was removed. Each stem segment was weighed and crushed in two volumes of PBS buffer. The resulting suspensions were subjected to a 10 fold serial dilution, and 0.1 mL aliquots were spread on agar plates containing NBY media. The number of CFU was determined after five d of incubation at 28 oC. Three replicate samples were taken and the experiment was conducted three times within each plant generation.
A Student t test was performed taking into account the number of CFU formed by the bacterial pathogens tested on each transgenic line at the different days of measurement.
Results and discussion
Several publications report that for bacterial diseases such as blight, a broad spectrum and higher resistance can be achieved by pyramiding more than two bacterial resistance genes into one line, compared with lines with a single resistance gene (Kim et al., 2009; Suh et al., 2009). An increasing attention has focused on the accumulation of major disease resistance genes in crop plants, although it is very difficult to pyramid genes using conventional breeding methods due to the dominance and epistasis effects of genes governing disease resistance (Suh et al., 2013). For that reason, a genetic engineering approach was chosen to pyramid the genes. To pyramid all three PRRs genes into one breeding line, the pCAMBIA-EFR:Ub:BAK1:Ub:FLS2 plasmid was constructed (Figure 1). The identity of the cassette was verified by restriction mapping and sequencing.
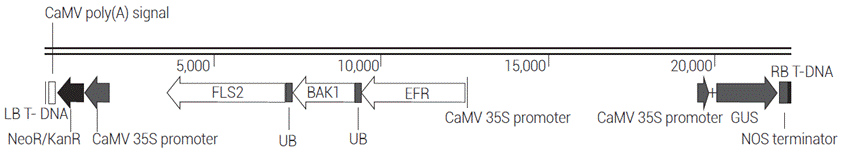
Figure 1 T-DNA region of the pCAMBIA-EFR:Ub:BAK1:Ub:FLS2 plasmid. The PRRs genes cassette was cloned into the plant expression vector pCAMBIA 2301. LB T-DNA: left border from nopaline C58 T-DNA; CaMV poly(A) signal: cauliflower mosaic virus polyadenylation signal; NeoR/KanR: aminoglycoside phosphotransferase gene from Tn5; CaMV 35S promoter: cauliflower mosaic virus 35S promoter; FLS2: FLAGELLIN-SENSING 2 gene from A. thaliana; UB: ubiquitin monomer; BAK1: BRI1-ASSOCIATED RECEPTOR KINASE gene from A. thaliana; EFR: EF-Tu RECEPTOR gene from A. thaliana; GUS: β-glucuronidase gene; NOS terminator: nopaline synthase terminator; RB T-DNA: right border from nopaline C58 T-DNA.
It is important to note that although there are previous reports of plants transformed with a PRR gene (Chinchilla et al., 2006; Lacombe et al., 2010), this is the first time that plants are transformed using a multigene cassette that includes two PRRs genes (AtEFR and AtFLS2) in addition to the BAK1 co-receptor gene (AtBAK1). This could have two advantages. On the one hand it could generate resistance to a broader spectrum of pathogenic species, including both gram-negative and gram-positive bacteria. And on the other hand the resistance generated could be stronger and more durable, but to be able to assert that it would be necessary to perform more assays to determine the contribution of the pyramiding effect on the resistance. It is a well-known fact that when the resistance of a plant to a certain pathogen relies on a single gene, it is rapidly overcome by the pathogen by selecting for mutants, recombinants, or immigrants that are better adapted to the resistant variety (Brown et al., 1997; Kumar and Nayak, 2010).
Tomato transformation is a well standardized routine procedure practiced in laboratory (Peña-Ramírez et al., 2007), and for the present study four evidences were used to verify the transgenic nature of the plants: GUS expression colorimetric assays, transgenes detection by PCR amplification using specific primers, detection of transcript levels of the transgenes by RT-PCR, and resistance against phytopathogenic bacterial infections as a result of the transgenes expression. It was possible to generate several transgenic tomato lines and four of these were selected that showed GUS activity in all the tissues analyzed (leaves and fruits) (Figure 2A).
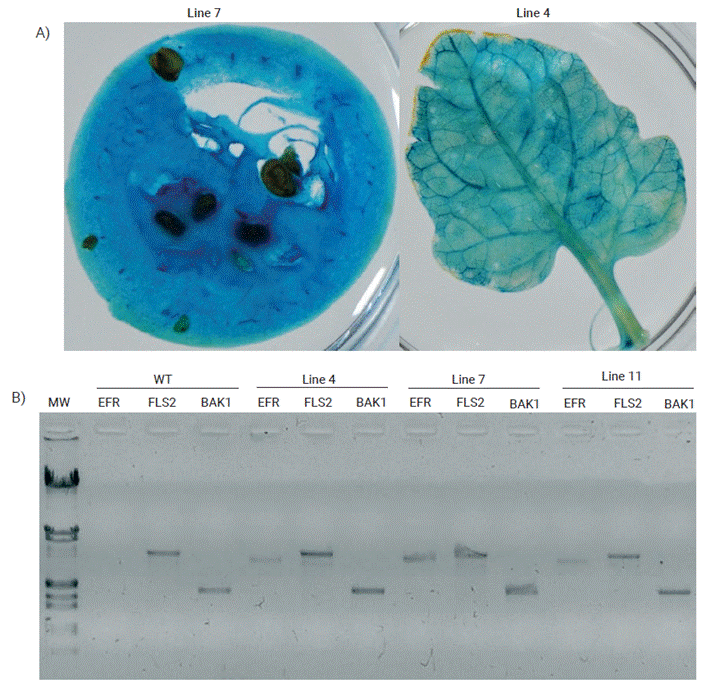
Figure 2 Molecular and histochemical evidence of the tomato transformation. (A) Stable GUS expression of F4 transgenic plants in fruit and leaf. (B) PCR of F4 transgenic and wild type (WT) plants with AtEFR, AtFLS2 and AtBAK1 primers. MW: Lambda DNA EcoRI/HindIII double digest as marker; EFR: 3,093 bp; FLS2: 3,519 bp; BAK1: 1,845 bp.
DNA was extracted from the four tomato lines and PCR was performed to determine whether the genes (AtEFR, AtBAK1 and AtFLS2) were present (Figure 2B). EFR gene amplified in the transgenic lines but not in the wild-type (WT) plants. This outcome was expected since tomato plants do not have an orthologue for such gene. However, FLS2 and BAK1 genes amplified not only in the transgenic lines but also in the WT, indicating that the tomato orthologues for those genes are similar enough to that from Arabidopsis, so they might be amplified using the same set of primers. The four tomato lines were allowed to self-pollinate to originate F1 to F4 generations. Within each generation GUS assays and PCR amplification of the genes were performed. Each consecutive generation showed an increasing percentage of positive GUS assays (85 % for F1, 91 % for F2, 94 % for F3 and 97 % for F4), which indicates that the transgenes were integrated into the plant genome and stably inherited, and that the tomato lines were trending towards homozygosity.
Plants were transformed with a single multi-cassette vector where the three genes were separated from each other by an ubiquitin monomer linker from tobacco and under the control of a common 35S promoter because this approach confers many advantages. First, the use of one promoter allows the coordinated and possible equimolar expression of the different proteins (Walker and Vierstra, 2007). Second, ubiquitin is rapidly and accurately processed in vivo by deubiquitinating enzymes, which release the attached proteins in free functional forms (Wilkinson, 2000). Third, this approach reduces the possibility of silencing the transgenes by using a single promoter, which might be a problem if more than one promoter is employed (Baulcombe, 2005).
It is important to notice that although PTI appears to be conserved across the plant kingdom, despite many similarities there are differences in the nature of PTI responses among different plant species (Nguyen et al., 2010). That is why it is feasible that transferring PRRs between plant families can confer new resistance capabilities against bacteria. In fact, it has already been proved that major resistance genes retain function when transferred between species. For example, the expression of EFR in the wild tobacco species Nicotiana benthamiana, that is normally unresponsive to EF-Tu, resulted in recognition of elf18 and activation of typical basal defenses (Zipfel et al., 2006). Furthermore, transgenic Solanaceae plants expressing EFR showed significant resistance to multiple gram-positive bacterial pathogens (Lacombe et al., 2010).
Flagellin, the building block of the eubacterial flagellum, is recognized by its cognate receptor FLS2 in nearly all plant species, but the recognized domain within flagellin is not necessarily the same for all plant species. For instance, members of both the Brassicaceae and Solanaceae families perceive flagellin; however, they vary in their sensitivity to different peptides of the protein (Robatzek et al., 2007). FLS2 receptors have been isolated from tomato, tobacco, and rice (Robatzek et al., 2007); all of these receptors display high levels of identity to Arabidopsis FLS2 at the amino acid level and also mediate flagellin perception; however, computational and phylogenetic approaches have suggested different amino acid residues as important for ligand binding (Albert et al., 2010; Boller and Felix, 2009). Considering the aforementioned, it was decided to express the AtFLS2 in tomato expecting that it would confer the tomato plants an extra flagellin epitope recognition site.
Challenging transgenic plants with phytopathogenic bacteria is the established method for testing the resistance or susceptibility of a given plant genotype to a specific pathovar (Zhao et al., 2003). It was decided to challenge plants with Pss, Pst and Cmm since they are important pathogens of tomato and the three of them are well characterized in the laboratory; more bacterial species and pathovars will be tested as they are available.
As expected, some of the transgenic lines showed resistance against the bacterial infections, even when the inoculum used for the challenges contained a much higher bacterial concentration than the one that can occur under natural infection. Line 7 in particular showed a reduction of near 60 and 73 % on colony forming units (CFU) when infected with Pss and Pst respectively compared with the WT plants by 10 d post-infection (dpi) (Figure 3A). Such differences in the number of CFU were also reflected in the general appearance of the plants since disease symptoms in plants from Line 7 were much less severe than those in WT plants (Figure 3B). Tomato is normally susceptible to infections by Pst since several isolates of the bacterium display amino acid polymorphisms within flagellins that mask their recognition in tomato (Cai et al., 2011).
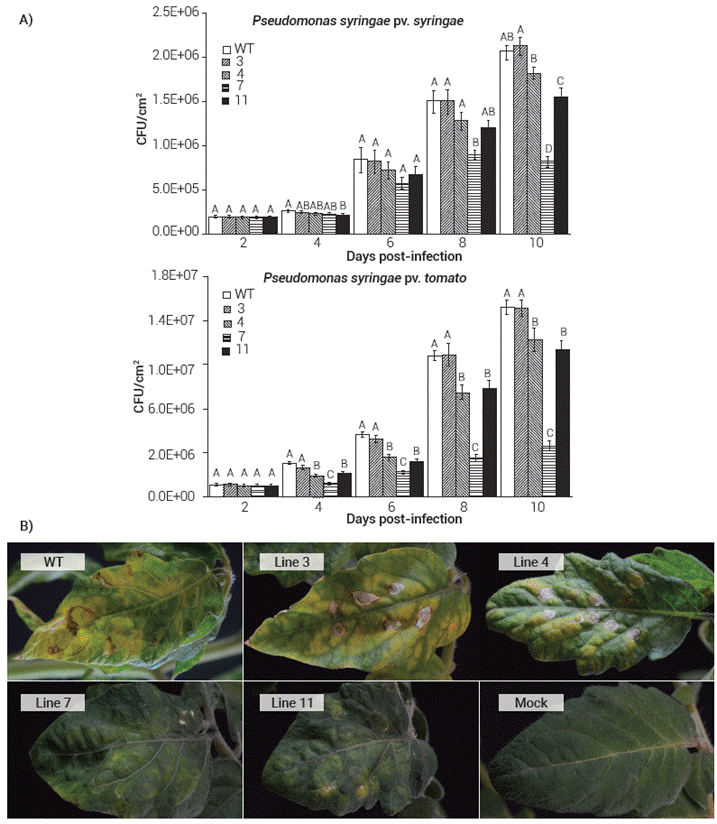
Figure 3 Transgenic expression of AtEFR, AtFLS2 and AtBAK1 in tomato confers resistance to P. syringae pv. syringae and P. syringae pv. tomato . (A) Colony forming units on the different F4 transgenic tomato lines and wild type (WT) plants after the bacterial infections. Results are average ± s.e.m.; means with the same letters are not significantly different (Student t, P ≤ 0.05). (B) Symptoms development on leafs of the different F4 transgenic tomato lines and wild type (WT) plants at 10 d post-infection with P. syringae pv. tomato. Mock represents a control treatment with no bacteria.
Previous studies had shown that AtFLS2 expression in tomato is sufficient to transfer the Arabidopsis flagellin perception system (Chinchilla et al., 2006), this is important since some studies have suggested that AtFLS2 has a broader recognition capacity than initially anticipated. For example, Ax21-derived peptides from Xanthomonas oryzae pv. oryzae activate FLS2-mediated Arabidopsis immunity, even when rice FLS2 does not appear to sense Ax21 or Ax21-derived peptides; that is, rice plants (Oryza sativa L.) lacking receptor XA21 (that still carry OsFLS2) do not respond to Ax21-derived peptides with measurable resistance (Danna et al., 2011). That may be a possible explanation to the observed resistance of the transgenic tomato lines against Pseudomonas in the present study, that heterologous expression of AtFLS2 activates plant immunity by recognition of another PAMP from Pseudomonas rather than flg22.
Additional evidence that supports these results is that Arabidopsis fls2 mutants have showed enhanced susceptibility to Pst DC3000 (Zipfel et al., 2006), demonstrating that AtFLS2 somehow perceives a PAMP from Pst activating the plant immunity. We hypothesize that for the resistance to Pseudomonas shown by the transgenic tomato plants in the present study, the contribution of AtFLS2 expression was more important than that from the expression of AtEFR, since it has already been reported that flagellin from P. syringae is more active triggering FLS-mediated PTI in Arabidopsis than flagellin from Agrobacterium tumefaciens or Sinorhizobium meliloti (Felix et al., 1999). Furthermore, EF-Tu from Pst DC3000 is much less active in eliciting PTI in Arabidopsis than EF-Tu from Agrobacterium (Kunze et al., 2004).
However, to be certain of this more studies and experiments are needed. It is important to notice that although the transgenic plants showed a reduced level of bacterial growth and disease symptoms, there was still a small but detectable degree of infection even in Line 7 which showed the best results. This indicates that the resistance conferred was not absolute and a possible explanation for this could be the presence of the AvrPto/AvrPtoB effectors in Pst DC3000, which are known to be strong suppressors of PTI (He et al., 2006).
In fact, it has been demonstrated that when delivered by Pst at natural levels, AvrPto and AvrPtoB target BAK1 and block the ligand-induced formation of PAMP-receptor complexes, thereby effectively impeding multiple PAMPsignaling initiation (Shan et al., 2008). Therefore, transforming tomato plants with an AtBAK1 gene, even when tomato contains an endogenous orthologue (Peng and Kaloshian, 2014), might results in overexpression of the BAK1 RLK which could probably counteract the attack of the AvrPto/AvrPtoB effectors and make the formation of PAMP-receptor complexes possible even in the presence of such effectors.
For the infections with Cmm the same pattern was observed despite using a much more concentrated inoculum than that a plant could encounter in nature, with some transgenic lines showing some resistance to the bacterium. Once again, the best results were found in plants from Line 7 which had a reduction of near 83 % on the CFU of Cmm compared with the WT tomato plants by 40 d postinfection (Figure 4A). The decrease in the number of CFU was also reflected on the general appearance of the plants, especially in Line 7 where the bacterial canker symptoms were much milder than those of WT plants (Figure 4B). Since Clavibacter is characterized by non-motility and therefore with no flagellar structures, it is tempting to hypothesize that the resistance shown by the transgenic tomato lines in the present study was not due to the heterologous expression of the AtFLS2, but rather to the expression of the AtEFR; but once again, more trials and experiments would be needed to confirm this.
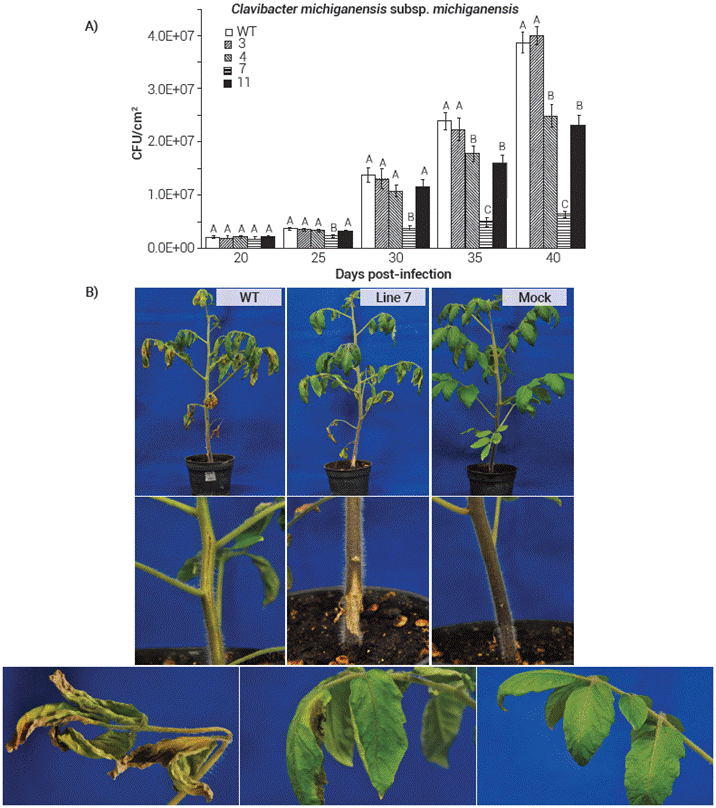
Figure 4 Transgenic expression of AtEFR, AtFLS2 and AtBAK1 in tomato confers resistance to C. michiganensis subsp. michiganensis . (A) Colony forming units on the different F4 transgenic tomato lines and wild type (WT) plants after the bacterial infections. Results are average ± s.e.m.; means with the same letters are not significantly different (Student t, P ≤ 0.05). (B) Bacterial canker symptoms development on the F4 tomato and wild type (WT) plants at 35 d post-infection with C. michiganensis subsp. michiganensis . Upper row: general view of the whole plant; middle row: close-up of the stem at the site of inoculation; bottom row: symptoms development on leaves. Mock represents a control treatment with no bacteria.
The PAMP recognized by the EFR receptor is the prokaryotic translation factor EF-Tu, the most abundant and one of the most conserved proteins in bacterial cells (Walters, 2015). Nevertheless, perception of the bacterial EF-Tu is found only in the Brassicaceae, all plants outside this family tested so far, including some Solanaceae, have failed to show the characteristic reactive oxygen species burst when challenged with the EF-Tu or its fully active elicitor peptide (elf18) (Zipfel et al., 2006). That means that expressing the EFR receptor in a Solanaceae (such as tomato in this case), confers the plant a novel tool for recognizing potential pathogens and triggering its basal immunity to fight them, as has been previously demonstrated by Lacombe et al. (2010).
In order to find an explanation to the difference on the resistance levels shown by the different transgenic lines, cDNA was synthetized from total RNA extracted from young leaves, and RT-PCR was performed on the cDNA to identify the transcript accumulation levels of the transgenes (Figure 5). Although transcript accumulation from FLS2 and BAK1 was detectable in the WT plants (because of tomato orthologues genes), Lines 4 and 11, but specially Line 7, showed higher transcript accumulation indicating an overexpression of those genes as a result of the heterologous expression of the Arabidopsis genes under a constitutive promoter.
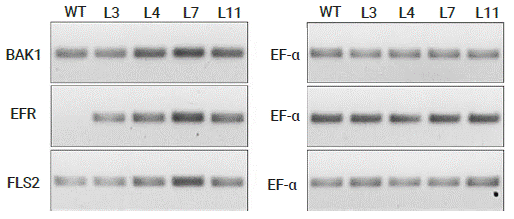
Figure 5 RT-PCR analysis of pCAMBIA-EFR:Ub:BAK1:Ub:FLS2 transgenic tomato lines using BAK1, EFR and FLS2 specific primers. A control RT-PCR of the house-keeping gene EF-α indicates equal loading.
Line 3 showed no higher transcript accumulation compared with the WT plants, thus explaining why plants from Line 3 showed little to none resistance to the bacterial infections. Nevertheless, Line 3 showed transcript accumulation of EFR (though only a little) indicating that plants from Line 3 were indeed transgenic but with no optimal expression of the transgenes. As it was expected, WT plants showed no transcript accumulation of EFR since that gene is restricted to Brassicaceae (Zipfel et al., 2006), while once again Line 7 showed the highest transcript accumulation, thus helping to explain why plants from such line had the lowest CFU counting and the mildest symptoms from the bacterial infections.
Conclusions
Tomato plants transformed with the specified construction are more resistant to infection by Pseudomonas syringae pv. syringae, P. syringae pv. tomato and Clavibacter michiganensis subsp. michiganensis than wild-type nontransformed plants. Whether the resistance generated by the heterologous expression of more than one gene related to plant innate immunity is stronger or more durable than that achieved by the expression of a single gene, it remains to be elucidated. This technology can potentially be applied to other important crops to confer resistance to a wide range of pathogenic bacteria.











 nueva página del texto (beta)
nueva página del texto (beta)


