Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista fitotecnia mexicana
versión impresa ISSN 0187-7380
Rev. fitotec. mex vol.39 no.1 Chapingo ene./mar. 2016
Artículo científico
Diversity of drought-responsive genes in central and peripheral populations of Trifolium purpureum Loisel
Diversidad de genes de respuesta a sequía en poblaciones centrales y periféricas de Trifolium purpureum Loisel
Ricardo Trejo-Calzada1*, Mary A. O'Connell2, Aurelio Pedroza-Sandoval1, Jesus G. Arreola-Ávila1 y Manuel Reveles-Hernández3
1 Unidad Regional Universitaria de Zonas Áridas, Universidad Autónoma Chapingo. 35230, Apartado Postal No. 8, Bermejillo, Durango, México. * Autor de correspondencia (rtrejo@chapingo.uruza.edu.mx)
2 College of Agricultural, Consumer and Environmental Sciences, New Mexico State University. 179 Gerald Thomas Hall. Las Cruces, New Mexico, USA.
3 Campo Experimental Zacatecas, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias. km 24.5 carr. Zacatecas-Fresnillo. 98001, Apartado Postal 196, Calera, Zacatecas.
Recibido: 24 de Febrero del 2015.
Aceptado: 3 de Noviembre del 2015.
Abstract
Drought-responsive genes may differ in structure and complexity in native populations of a species established in different ecosystems. Peripheral populations may be a source of genetic variability for breeding cultivated plants for abiotic stresses tolerance and the target for core collections in germplasm preservation programs. Genetic studies including both peripheral and central populations are still limited. This research evaluated genetic diversity of drought-responsive genes in peripheral and central populations of Trifolium purpureum Loisel. Genomic DNA isolated from leaves of three northern and three southern populations of Israel was digested with restriction enzymes and hybridized with four drought-induced and four drought-repressed gene fragments. RFLPs were analyzed for gene diversity, molecular variation and fixation indexes (FST). Gene diversity of drought-induced genes ranged from 0.1 to 0.42 but differences of individual or pooled genes between central and peripheral populations were nonsignificant. Gene diversity for drought-repressed genes ranged from 0.08 to 0.348. Even though there were no differences for individual genes, a joint analysis showed a significantly larger (P ≤ 0.05) gene diversity in peripheral populations of T. Purpureum than in central ones. Variation within populations for both drought-induced and drought-repressed genes was the main component of molecular variance. Fixation index (FST) for drought-induced genes was between 0.029 and 0.214 while for drought repressed genes it was between 0.04 and 0.33. Results of this study show that peripheral population might be a reservoir for drought-responsive genes.
Key words: Trifolium purpureum, abiotic stress, adaptation, arid lands, gene flow, molecular variation.
Resumen
Los genes de respuesta a sequía pueden diferir en estructura y complejidad en poblaciones nativas de una especie establecidas en diferentes ecosistemas. Las poblaciones periféricas pueden constituir una fuente de variabilidad para el mejoramiento de plantas cultivadas para tolerancia a estreses abióticos, y también pueden ser el objetivo para colecciones núcleo en programas de conservación de germoplasma. Al respecto, estudios genéticos que incluyan tanto poblaciones periféricas como centrales son escasos. El objetivo de este estudio fue evaluar la diversidad genética de genes de respuesta a sequía en poblaciones centrales y periféricas de Trifolium purpureum Loisel. El ADN genómico aislado de hojas de tres poblaciones del norte y tres poblaciones del sur de Israel fue digerido con enzimas de restricción e hibridados con cuatro fragmentos de genes inducidos y cuatro de genes reprimidos por sequía. Los polimorfismos de longitud de fragmentos de restricción (RFLP) fueron analizados para diversidad genética, variación molecular e índices de fijación (FST). La diversidad génica de los genes inducidos por sequía varió de 0.1 a 0.42 pero no hubo diferencias significativas para los genes individuales o para los grupos de genes entre las poblaciones periféricas y centrales. La diversidad génica para genes reprimidos por sequía tuvo valores entre 0.08 y 0.348. Aun cuando no hubo diferencias para genes individuales, un análisis conjunto mostró una diversidad genética significativamente mayor (P ≤ 0.05) en poblaciones periféricas que en poblaciones centrales de T. purpureum. La variación dentro de poblaciones fue el principal componente de variación molecular tanto para genes inducidos como reprimidos por sequía. El índice de fijación (FST) para genes inducidos por sequía varió entre 0.029 y 0.214, en tanto que para genes reprimidos por sequía estuvo entre 0.04 y 0.33. Los resultados de este estudio muestran que las poblaciones periféricas pueden ser un reservorio de genes de respuesta a sequía.
Palabras clave: Trifolium purpureum, adaptación, estrés abiótico, flujo de genes, variación molecular, zonas áridas.
INTRODUCTION
Plant adaptation to the environment involves a wide range of development strategies, morphological features, biochemical mechanisms, and physiological traits under genetic control (Yamaguchi-Shinozaki and Shinozaki, 2006). Genes involved in the responses of plants to short-term and season-long water deficit have been studied in great detail in many agronomic plants but in much less detail in wild species. Studies in some cultivated plants have shown that there is a number of genes and gene families expressed under drought stress (Wang et al., 2011). These drought-induced genes can be modeled to be involved in morphological, physiological and molecular features that make the plants able to deal with water deficit (Shinozaki and Yamaguchi-Shinozaki, 2007).
Drought-responsive gene families may differ in structure and complexity in native populations of a species established in different ecosystems. The central-marginal model in evolutionary ecology proposes that populations near the center of the range (central) of a species are highly dense, and show high levels of phenotypic and genetic variation, while populations on the edge of the range (peripheral) are isolated, bare, and chromosomally monomorphic (Brussard, 1984).
The center of the range of a species will coincide with the geographic region that is the most ecologically favorable. The periphery of a range will usually correspond to an area of extreme marginality for the species (Vucetich and Waite, 2003). Thus, peripheral populations are expected to be genetically distinct from central populations since they may face different selection conditions. The differences between central and marginal populations will depend on the differences of the effective population size and the gene flow (Eckert et al., 2008).
Peripheral populations may constitute a source of genetic variability for breeding cultivated plants for tolerance to abiotic stresses. They may also be the target for constructing core collections in germplasm preservation programs. However, genetic studies including both peripheral and central populations are still limited (Eckert et al., 2008). Nowadays, peripheral populations are acquiring importance for gene conservation, because they may possess genotypes of future adaptive potential especially under climate change conditions (Pandey and Rajora, 2012a).
Purple clover (Trifolium purpureum Loisel) is a species native to Mediterranean regions of Europe. This species is now found as natural populations in regions with environments more hostile than those from where it was originated. Therefore, some T. purpureum populations can be called central populations, as they are located in favorable environments. Other populations may be called peripheral as they populate unfavorable environments. The presence of this species in grasslands of temperate and semiarid regions may involve differences in diversity of drought-responsive genes among populations.
In this study, several drought-responsive genes were used to perform RFLP analyses on central and peripheral populations of T. purpureum to evaluate genetic diversity of drought responsive genes.
MATERIALS AND METHODS
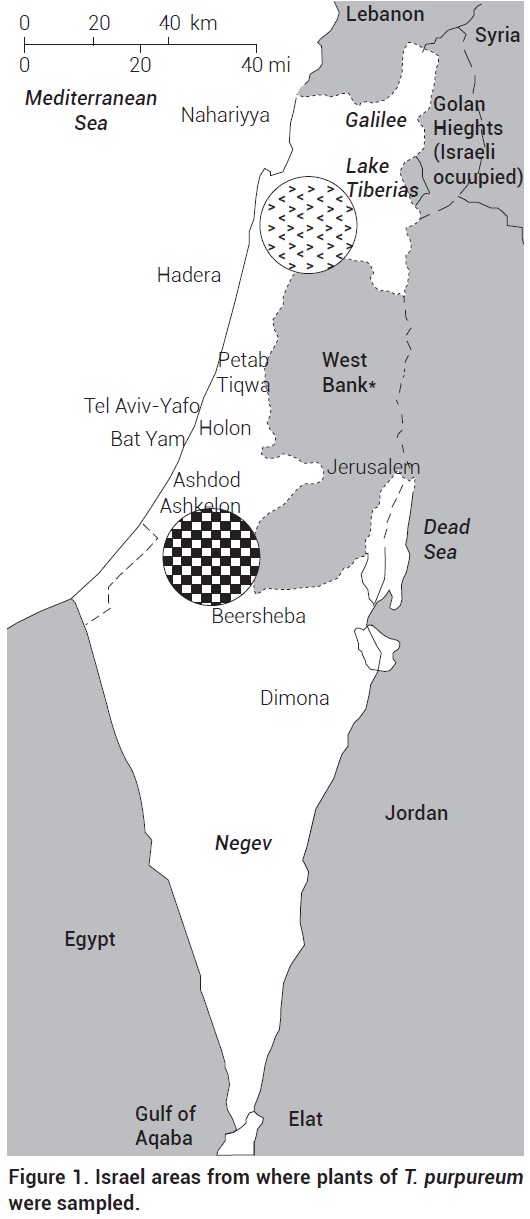
T. purpureum Loisel seeds from different locations of Northern [(Givat-Hamore (32° 37' 41.05" N, 35° 19' 56.12" E), Carmel (32° 43' 44.39" N, 35° 2' 45.07" E), Kefar-Ha-choresh (32° 42' 12.08" N, 35° 16' 20.27" E) and Gilboa (32° 25' 56.76" N, 35° 24' 54.27" E)] and Southern [(Pura (31° 29' 47.47" N, 34° 46' 28.52" E), Lalow, Sausana, Beeri (31° 26' 17.31" N 34° 29' 15.19" E), and Tel Gome (31° 23' 5.22" N, 34° 27' 7.98" E)] Israel were collected (Figure 1). The Northern region from where one group of the seeds was collected has a Mediterranean climate with annual winter rain of 450-600 mm. The Southern region, from where the other seed group was collected, is located in the outer edge of the extreme Judean desert with ~300 mm of annual rain. Seeds were grouped as central (northern located) or peripheral (southern located) according to their origin. Those seeds were used to grow plants in a greenhouse at New Mexico State University, USA.
Drought-responsive cDNA clones from T. purpureum were isolated from a cDNA library made from leaves of drought-stressed plants. Drought-induced and drought-repressed cDNA clones were confirmed by Northern analysis and later sequenced. Eight drought-responsive cDNAs were used as probes for the analysis of gene diversity of T. purpureum populations. Five out of the eight cDNA sequences were submitted to ESTs GenBank database.
A Puregene© kit was used to isolate DNA from leaves. Samples of leaf tissue were taken from at least 20 plants from each of three northern and three southern populations (n = 120). A total of 10 μg DNA was restricted with 2 μl of EcoRI or 2 ul of HindIII. The digested samples were electrophoresed in a 0.8 % agarose (Type 1 low EEO) gel at 70 V (4 V/cm). A total of 20 DNA samples from each population were set in a single gel (Figure 2A). Four replica gels were made for each population. DNA was transferred onto Zetabind nylon membranes using alkaline solution (0.6 M NaCl and 0.4 M NaOH) and dried in a vacuum oven at 80 °C for 1.5 to 2 h. The membranes were prehybridized overnight in 0.5 M Na2HPO4, 1 mM EDTA, 1 % BSA, 7 % SDS, and pH 7.2 solution. Those membranes were then hybridized overnight with [32P] dCTP labeled cDNA probes at 68 oC and shaking conditions. After hybridization, membranes were washed with 0.5 M Na2HPO4, 0.5 M EDTA, 10 % SDS, 3 % fish guts, and pH 7.2 solution. The membranes were exposed to x-ray film Midwest Scientific at -80 oC for varying periods of time (24 hours to 7 days) (Figure 2B).
The polymorphisms for each of the probes were identified on autoradiograms and scored as binary data (Figure 2B). The matrices generated were analyzed with Popgene, version 1.31software (Yeh and Boyle, 1997) to estimate gene diversity, genetic distance, and genetic differentiation (Nei and Kumar, 2000).
An analysis of molecular variance (AMOVA) was performed taking into account the frequencies of haplotypes generated by each probe. The AMOVA, F-statistics and gene flow [Nm = ((1-FST)-1))/4] were estimated using the software Arlequin, version 3.0 (Souza et al. 2002; Excoffier et al., 2005).
RESULTS AND DISCUSSION
Drought-induced genes showed great complexity as measured by the number of different fragments in the hybridized Southern blots of T. purpureum. Several polymorphic loci were identified by using drought-induced cDNAs as probes. The number of polymorphic loci produced by the hybridization of southern blots containing genomic DNA of T. purpureum varied between 5 and 14 (Table 1).
Drought-repressed genes showed similar results to drought-induced genes regarding the number of loci and the complexity of the genes. Drought-repressed genes produced 6 to 11 loci when used to hybridize Southern blots containing genomic DNA from T. purpureum plants (Table 1).
The number of polymorphic loci may vary depending on the species and markers used. Thus, Coulibaly et al. (2002) found 114 polymorphic loci into 106 accessions of Vigna unguiculata L. by AFLP markers, while Sebastian et al. (2010) using RAPD markers found 3 to14 polymorphic bands per primer in Tylophora rotundifolia.
The amount of genetic variability of a population is a function of the genetic diversity originally available to the species and of the later influence of processes such as selection, gene flow, and the mating system (Despres et al., 2002). In this study, the highest values of gene diversity for the dehydrin related protein gene and the probable transcription factor gene were observed in central populations with H = 0.2857 and H = 0.4407, respectively. However, the highest values of gene diversity for the LEA-like protein gene and the arginine decarboxyla-se gene were observed in peripheral populations with H = 0.3817 and H = 0.369, respectively.
The differences of average gene diversity between central and peripheral populations were not significant for any of the drought-induced genes used as probes. Khanlou et al. (2011) using AFLP markers in three cultivars of T. re-pens measured genetic diversity values of 0.319, 0.289 and 0.272 for different cultivars, which are similar to the ones found in this study.
The AMOVA showed that variation within populations was the main component of the variance for the drought-induced genes. Around 88 % of the variation in drought-induced genes was found within populations. In addition, variation among populations within groups was highly variable in the four drought-induced genes. The difference of variance among populations within groups was significant for all the genes, except for the one that codes for arginine decarboxylase. In contrast, variation among groups of all drought-induced genes was very low and not significant (Table 2).
The estimated values of gene flow for drought-induced genes were high with a relatively wide range. Gene flow ranged from Nm = 0.91 for LEA-like protein gene to Nm = 8.3 for dehydrin related protein gene. The average gene flow for the group of drought-induced genes was 3.4. After using nuclear SSR and cpSSR markers Perderau et al. (2014) found high Nm values (2.29 and 2.76, respectively) of gene flow in populations of Salix caprea. Also, Welt et al. (2015) found high levels of gene flow (3.08 and 4.44) in Brassica rapa in two different years (1997 and 2004) by using microsatellite analysis. Fixation index (FST) showed that in three out of the four drought-induced genes there were significant differences among populations, with values ranging from 0.029 to 0.214 (Table 3).

The fixation index FST is a suitable measure of genetic differentiation among subpopulations (populations within groups). This index measures the overall reduction in average heterozygosity in subpopulations regarding the total heterozygosity, and is the most inclusive measure of population substructure. FST has a theoretical minimum of 0 (when there is not genetic divergence) and a maximum of 1 (when alternative alleles are fixed in different subpopulations). Thus, FST is used as a relative measure of population structure and a comparative estimation of gene flow (Miller et al., 2008).
Gene diversity of drought-induced genes in central and peripheral populations of T. purpureum was larger than that reported for T. pratense and other out crossing species. The expected heterozygosity of 12 allozymes in 9 populations of T. pratense from Southeastern United States was H = 0.250. Some widely distributed species have shown an average gene diversity of H = 0.202 (Hagen and Hamrick, 1998).
The relatively high gene diversity for drought-induced genes observed in T. purpureum could be attributed to the cross pollination mating system, its annual growth habit, and the fast growth of populations. Cross pollinated species tend to have higher levels of variability within populations but a smaller degree of differentiation among populations than selfing species. Moreover, genetic differentiation is greater in annuals than in perennials (Fisher et al., 2000). Studies in Calystegia collina showed high values of gene diversity, where more than 40 % of the genetic variation was due to variation among populations; this high gene diversity was attributed to self-incompatibility, vegetative reproduction, and possible mutations in long-lived clones (Wolf et al., 2000).
Gene diversity for drought-repressed genes ranged from 0.08 to 0.348. However, average gene diversity was not significantly different in central or peripheral populations for individual drought-repressed genes. Gene differentiation among populations, measured as fixation index (FST), was significant for all drought-repressed genes in T. purpureum. The highest differentiation was for the gene that codes for a nucleotide binding protein with FST ~0.33, while the lowest differentiation was produced by the gene coding for ATP synthase with FST value ~0.04. Also, the estimated gene flow was high for all four drought-repressed genes (Table 4).

The AMOVA identified that variation within populations of T. purpureum was the most important component of the total variation (~67 to ~96 %) of drought-repressed genes. The average contribution of variability within populations to total genetic variability of drought-repressed genes was ~85 %. Variation among populations was significant, and on the average it accounted for ~13.5 % of the total genetic variance. Variation among groups (central vs. peripheral) was low and not significant for individual drought-repressed genes (Table 5).
A pooled analysis of the four drought-repressed genes showed that genetic diversity for the group of genes was significantly larger (P ≤ 0.05) in peripheral populations than in central populations of T. purpureum. Hagen and Hamrick (1998) found that populations of T. pratense from northern and southern USA were weakly differentiated by genetic diversity. The total genetic variation due to differences among regions and among populations was 0.013 and 0.049, respectively; most of this genetic diversity occurred within populations (93.8 %).
The main component of variation of drought-responsive genes in central and peripheral populations of T. purpu-reum was the variance within populations. The high variation within populations may be explained by the mating system of the species and their wide distribution. In general, genetic diversity for drought-responsive genes in T purpureum was high. On the contrary, in some species a very low genetic diversity has been found. In populations of wild rice (Oryza granulata) the gene diversity was in the range of 0.0 to 0.029. The low gene diversity was attributed to geographically narrow distribution of the populations as a colonizing species (Gao et al., 2000).
Some studies show a higher genetic differentiation in peripheral populations than in central populations, such as those on eastern white cedar (Thuja occidentalis L.) (Pandey and Rajora, 2012b). Findings by Cortés et al. (2012) show that gene diversity for genes associated to drought may be significantly different in contrasting populations. ABA stress response genes Asr1 and Asr2 had different gene diversity in wild and cultivated populations of Phaseolus vulgaris L. Also, Trejo-Calzada and O'Connell (2005) showed that populations of Dactylis glomerata from an arid region of Israel had a greater genetic diversity for drought-responsive genes (0.388, repressed; 0.340, induced) than populations of plants collected in the northern Mediterranean region of Israel (0.308, repressed; 0.314, induced).
CONCLUSIONS
Genetic diversity for both drought-induced and drought-repressed genes was high; however, the differences in average gene diversity between central and peripheral populations were not significant for any of the drought-responsive genes. Variation within populations was the main component of the variance of drought-induced (88 %) and drought-repressed genes (85 %). On the contrary, variation among groups for all individual drought-induced and drought-repressed genes was very low and not significant. Gene differentiation or fixation index (FST) was significant for all the genes used as probes in this study, except for the one that code for arginine decarboxylase. The estimated gene flow was relatively high for drought-induced and drought-repressed genes.
BIBLIOGRAPHY
Brussard P. F. (1984) Geographic patterns and environmental gradients: The Central-Marginal model in Drosophila revisited. Annual Review of Ecology and Systematics 15:25-64. [ Links ]
Cortés A. J., M. C. Chavarro, S. Madriñán, D. This and M. W. Blair (2012) Molecular ecology and selection in the drought-related Asr gene polymorphisms in wild and cultivated common bean (Phaseolus vulgaris L.). BMC Genetics 13:58. [ Links ]
Coulibaly S., R. S. Pasquet, R. Papa and P. Gepts (2002) AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theoretical and Applied Genetics 104:358-366. [ Links ]
Despres, L., S. Loriot and M. Gaudeul (2002) Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Molecular Ecology 11:2337-2347. [ Links ]
Eckert C. G., K. E. Samis and S. C. Lougheed (2008) Genetic variation across species' geographical ranges: the central-marginal hypothesis and beyond. Molecular Ecology 17:1170-1188. [ Links ]
Excoffier L., G. Laval and S. Schneider (2005) Arlequin (version 3.0): n integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47-50. [ Links ]
Fisher M., R. Husi, D. Prati, M. Peintinger, M. van Kleunen and B. Schmid (2000) RAPD variation among and within small and large populations of the rare clonal plant Ranunculus reptans (Ranunculaceae). American Journal of Botany 87:1128-1137. [ Links ]
Gao L. Z., S. Ge and D. Y. Hong (2000) Low levels of genetic diversity within populations and high differentiation among populations of a wild rice, Oryza granulata Nees et Arn. ex Watt., from China. International Journal of Plant Science 161:691-697. [ Links ]
Hagen M. J. and J. L. Hamrick (1998) Genetic variation and population genetic structure in Trifolium pratense. Journal of Heredity 89:178-181. [ Links ]
Khanlou K. M., K. Vandepitte, L. Kheibarshekan and E. Van Bockstaele (2011) Towards an optimal sampling strategy for assessing genetic variation within and among white clover (Trifolium repens L.) cultivars using AFLP. Genetics and Molecular Biology 34:252-258. [ Links ]
Miller J. R., B. P. Wood and M. B. Hamilton (2008) FST and QST under neutrality. Genetics 180: 1023-1037. [ Links ]
Nei M. and S. Kumar (2000) Molecular Evolution and Phylogenetics. Oxford University Press. New York, USA. 333 p. [ Links ]
Pandey M. and O. P. Rajora (2012a) Genetic diversity and differentiation of core vs. peripheral populations of eastern white cedar, Thuja occidentalis (Cupressaceae). American Journal of Botany 99:690-699. [ Links ]
Pandey M. and O. P. Rajora (2012b) Higher fine-scale genetic structure in peripheral than in core populations of a long-lived and mixed-mating conifer-eastern white cedar (Thuja occidentalis L.). BMC Evolutionary Biology 12:48. [ Links ]
Perderau A. C., C. T. Kelleher, G. C. Douglas and T. R. Hodkinson (2014) High levels of gene flow and genetic diversity in Irish populations of Salix caprea L. inferred from chloroplast and nuclear SSR markers. BMC Plant Biology 14:202. [ Links ]
Sebastian V. A., L. D. Cruz, R. B. Subramanian and V. J. Braganza (2010) Assessment of genetic diversity within and among populations of Tylophora rotundifolia using RAPD markers. Gene Conserve 9:94-117. [ Links ]
Shinozaki K. and K. Yamaguchi-Shinozaki (2007) Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58:221-227. [ Links ]
Souza F.L., A. F. Cunha, M. A. Oliveira, G. A. G. Pereira and S. F. dos Reis (2002) Estimating dispersal and gene flow in the neotropical freshwater turtle Hydromedusa maximiliani (Chelidae) by combining ecological and genetic methods. Genetics and Molecular Biology 25:151-155. [ Links ]
Trejo-Calzada R. and M. A. O'Connell (2005) Genetic diversity of drought-responsive genes in populations of the desert forage Dactylis glomerata. Plant Science 168:1327-1335. [ Links ]
Vucetich J. A. and T. A. Waite (2003) Spatial patterns of demography and genetic processes across the species' range: null hypotheses for landscape conservation genetics. Conservation Genetics 4:639-645. [ Links ]
Wang D., Y. Pan, X. Zhao, L. Zhu, B. Fu and Z. Li (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12:149. [ Links ]
Welt R. S., A. Litt and S. J. Franks (2015) Analysis of population genetic structure and gene flow in an annual plant before and after a rapid evolutionary response to drought. AoB PLANTS Advance Access. http://aobpla.oxfordjournals.org/content/early/2015/03/27/aobpla.plv026.full.pdf+html DOI:10.1093/aobpla/plv026. [ Links ]
Wolf A. T., R. H. Hower and J. L. Hamrick (2000) Genetic diversity and population structure of the serpentine endemic Calystegia collina (Convolvulaceae) in Northern California. American Journal of Botany 87:1138-1146. [ Links ]
Yamaguchi-Shinozaki K. and K. Shinozaki (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57:781-803. [ Links ]
Yeh F. C. and T. J. Boyle (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belgian Journal of Botany 129:157-163. [ Links ]














