Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista fitotecnia mexicana
versión impresa ISSN 0187-7380
Rev. fitotec. mex vol.34 no.4 Chapingo oct./dic. 2011
Artículos científicos
Genetic diversity and differentiation of Pseudotsuga menziesii (Mirb.) Franco populations in México
Diversidad genética y diferenciación de las poblaciones de Pseudotsuga menziesii (Mirb.) Franco en México
Jorge Cruz-Nicolás, J. Jesús Vargas-Hernández *, Porfirio Ramírez-Vallejo and Javier López-Upton
Colegio de Postgraduados-Campus Montecillo. Km. 36.5 Carretera México-Texcoco. 56230, Montecillo, Texcoco, Estado de México, México.*Author for correspondence (vargashj@colpos.mx)
Recibido: 23 de Abril del 2010.
Aceptado: 03 de Noviembre del 2011.
Abstract
Mexican populations of Douglas-fir [Pseudotsuga menziesii (Mirb.) Franco] constitute valuable gene pools for conservation and breeding programs of this species. Genetic diversity and population divergence were estimated using 18 isozyme loci and samples from 11 natural Douglas-fir populations in México. Genetic diversity was high at the species level (83.3 % polymorphic loci and 2.9 alleles per locus), but low at the population level (28.3 % polymorphic loci and 1.52 alleles per locus), particularly for populations from central México. A high level of population differentiation was found (Fst = 0.298), indicating that each population shares only a small fraction of the genetic diversity within the species and that genetic drift might have an important role in shaping the structure of genetic diversity in these populations. Genetic distances among populations in the Sierra Madre Oriental were positively correlated with geographic distances (r = 0.849), but the association was not significant according to the Mantel test. Populations from northeastern México separated from all others, whereas population Mohinora from northwestern México, joined those from central México. These results emphasize the priority for in situ conservation of Douglas-fir populations in central México.
Key words: Pseudotsuga menziessii, fragmentation, genetic drift, genetic structure, in situ conservation, population differentiation.
Resumen
Las poblaciones mexicanas de Pseudotsuga menziesii (Mirb.) Franco contienen acervos génicos valiosos para los programas de conservación y mejoramiento genético. Se estimó la diversidad genética y la divergencia de poblaciones utilizando 18 loci isoenzimáticos en muestras de 11 poblaciones naturales de P. menziesii en México. Se encontró una amplia diversidad genética a nivel de la especie (83.3 % loci polimóricos y 2.9 alelos por locus), pero reducida a nivel de población (28.3 % loci polimórficos y 1.52 alelos por locus), especialmente en las poblaciones de la región central de México. Se encontró una alta diferenciación genética entre las poblaciones (Fst = 0.298), lo que indica que cada población representa una pequeña fracción de la diversidad genética de la especie y que la deriva genética podría tener un papel importante en moldear la estructura de la diversidad genética en estas poblaciones. Las distancias genéticas entre las poblaciones de la Sierra Madre Oriental se correlacionaron positivamente (r = 0.849) con las distancias geográficas entre ellas, pero la asociación no fue significativa de acuerdo con la prueba de Mantel. Las poblaciones del noreste de México se diferenciaron del resto, mientras que la población de Mohinora en el Noroeste de México se agrupó con las del centro del país. Estos resultados enfatizan la prioridad de la conservación in situ de las poblaciones en el centro de México.
Palabras clave: Pseudotsuga menziesii, fragmentación, deriva genética, estructura genética, conservación in situ, diferenciación de poblaciones.
INTRODUCTION
Douglas-fir [Pseudotsuga menziesii (Mirb.) Franco] is one of the most economically important timber species in the world (Hermann and Lavender, 1999). Because of its economical and ecological value in North America, this species has been thoroughly studied in United States and Canada, where it has been the focus of several domestication programs for the last 50 years (Howe et al., 2006). The natural range of Douglas-fir spreads down into México, where it is a component of the mosaic of forest ecosystems found across the mountainous landscapes. Even though locally this species is not economically important for timber production, the genes residing in these populations might become important for domestication programs elsewhere and for long-term conservation of genetic resources, particularly in the face of climate change. They constitute an unique component of biodiversity in México, particularly within the temperate forest communities (Domínguez, 1994), so maintaining its habitat is also crucial for conserving all the associated species.
Douglas-fir is distributed along three mountain ranges in México (Martínez, 1963; Debreczy and Racz, 1995): Sierra Madre Occidental in the northwest, Sierra Madre Oriental in the northeast and central México, and Sierra Madre del Sur in the south. In contrast with its continuous distribution in Canada and United States, natural populations of Douglas-fir in México are small and isolated, commonly mixed with other conifer and hardwood species (Domínguez, 1994), thus having an extremely fragmented and discontinuous distribution. In the past, a controversy about the number of Pseudotsuga species growing in México was raised (Martínez, 1963; Little, 1979), but recent studies suggest that all Mexican Douglas-fir populations belong to the taxon Pseudotsuga menziesii (Debreczy and Racz, 1995; Gernandt and Liston, 1999). The existence of a broad phenotypic variation among populations has been reported for morphological traits (Reyes-Hernández et al., 2006), shoot phenology (Acevedo-Rodríguez et al., 2006) and seedlings growth traits (Juárez-Agis et al., 2006). Despite the importance of these populations as valuable pools of genetic resources for breeding programs, their genetic structure and level of diversity are still unknown.
Determining the genetic diversity within a species and its distribution pattern among and within populations is important for the conservation and management of genetic resources. This information allows establishing priorities for conservation and, at the same time, setting up the most appropriate sampling strategy for using and maintaining this genetic diversity. When most genetic diversity resides within populations and there is no significant differentiation, conserving a few of them would be enough to save the gene pool within the species (Brown and Hardner, 2000); however, when the genetic diversity has a more complex structure and populations become differentiated, the conservation strategy becomes more complicated.
In this study, data on the level and structure of genetic diversity in populations of Mexican Douglas-fir are presented. This information could be useful to set up appropriate measures for conservation of the genetic resources in these populations, particularly those from central México which are under strong human and environmental pressures. The specific objectives of the study were a) to determine the magnitude and structure of genetic diversity in the Mexican populations of Douglas-fir, focusing on the level of differentiation within the central region of México; and b) to evaluate whether the genetic differentiation among populations follows a particular geographic pattern.
MATERIALS AND METHODS
Populations sampled
Seed samples from a total of 170 mother trees from 11 natural populations of Douglas-fir distributed throughout its natural range in México (Figure 1) were included in the study. The geographic coordinates and number of trees sampled for each population are given in Table 1. A minimum distance of 50 m among mother trees was maintained to reduce the possibility of relatedness; seed lots were kept separated and identified by mother tree. Number of sampled trees per population varied because of differences in population size, seed availability and germination capacity (Mápula-Larreta et al., 2007).
Tissue extraction and electrophoresis procedure
Seed was first imbibed in a 1% H2O2 solution during 24 h, stratified at 4 °C for 50 d (Mápula-Larreta et al., 2008), and then germinated in Petri dishes with humid paper towels within a germination chamber at 24 °C. The haploid mega-gametophyte tissue was extracted from each seed when the radicle was about 5 mm long, and macerated in an Eppendorf tube, adding 100 (iL of extraction buffer (USDA Forest Service, 2003). A sample of six to seven seeds per tree was used to infer on the genotype of mother trees.
Electrophoresis runs followed the protocols of the National Forest Genetics Electrophoresis Laboratory (NFGEL) of the USDA Forest Service (USDA Forest Service, 2003). Eleven isozyme systems for which polymorphism had already been shown for Douglas-fir in previous studies (Li and Adams, 1989; Adams et al., 1990) were evaluated, including diaphorase (DIA), esterase (EST), glutamate dehydrogenase (GDH), glutamate oxaloacetate-transaminase (GOT), isocitrate dehydrogenase (IDH), leucine amino peptidase (LAP), malate dehydrogenase (MDH), malic enzyme (ME), phosphoglucose isomerase (PGI), phosphoglucomutase (PGM), and 6-phosphogluconate dehydrogenase (6-PGD). For DIA and GOT, the staining procedure of Stuber et al. (1988) was used.
A total of 18 loci were scored; samples of mega-gametophyte tissue from Pinus resinosa Ait., a species completely monomorphic for these loci, were included as control in each gel. For those enzymes in which several activity zones were detected, loci were identified with the same enzyme code followed by successive numerals, starting by the fastest migrating zone. Allele designation was done considering for each locus the most frequent allele in the whole sample as number 1, and those less frequent were numbered successively according to their relative distance from the origin.
Estimation of genetic diversity and genetic structure parameters
Parameters of genetic diversity, including gene frequency, number of alleles per locus and percentage of polymorphic loci (frequency of most common allele < 0.95) were calculated for each population, following the procedures of Brown and Weir (1983); in addition, observed (Ho) and expected (He) heterozygosity were estimated according to Nei (1978), corrected for small samples. To determine the genetic structure and extent of population differentiation, the F-statistics (Fit, Fis, and Fst) were estimated using BIOSYS-1 (Swofford and Selander, 1989). F¡t and F represent the deviations from expected heterozigosity in overall and within populations, respectively, with positive values indicating a deficiency of heterozygotes, and negative values an excess; Fst indicates the degree of genetic differentiation among populations. Genetic flow was estimated as the number of migrants per generation (Nm) with the procedure described by Slatkin and Barton (1989), using the following equation:
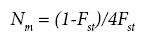
Genetic distances between populations were estimated according to Nei (1978), and geographic distances were calculated based on the geographic coordinates of populations (Byers, 1997) obtained with a GPS instrument during the field sampling. The significance of the correlation between genetic and geographic distances was tested with the Mantel test (Jørgensen et al., 2002). The genetic distances matrix was also used for grouping populations with the UPGMA method, using PHYLIP 3.6 (Felsenstein, 1995); to test the consistency of grouping, 500 bootstrap samples of gene frequencies for 15 polymorphic loci were generated using the same software.
All genetic analyses were done first including all 11 populations sampled and then repeated after excluding those populations (4) with a limited number of mother trees sampled. However, since there were no major differences between the results of both analyses, the interpretation was based primarily on the whole data set.
RESULTS AND DISCUSSION
Genetic diversity in Mexican Douglas-fir populations
Overall, 22 loci were resolved for the 11 isozyme systems assayed; but four of them (PGI-1, MDH-3, DIA-1 and 6-PGD1) were excluded from further analysis because they were inconsistently stained; three of the remaining 18 loci were completely fixed across populations, and 15 were polymorphic in at least one population, so 83.3% of the loci were polymorphic at the species level. A total of 44 alleles were identified in the 15 polymorphic loci, for an average of about three alleles per locus; thus, Mexican Douglas-fir is genetically diverse globally, as most coniferous species elsewhere (Hamrick and Godt, 1989). At the population level the genetic diversity was much lower, with an average of 1.5 alleles per locus and only 28.3 % of polymorphic loci. Moreover, a wide variation was found among populations for these genetic parameters (Table 2). On average, genetic diversity in these Mexican Douglas-fir populations seems to be lower than in populations from other regions in North America (Merkle and Adams, 1987; Li and Adams, 1989; El-Kassaby and Ritland, 1996).
Populations from northern México showed higher polymorphism than those from central México (Table 2), which are located at the margins of the species distribution and have a higher degree of isolation and fragmentation (Domínguez, 1994; Mapula-Larreta et al., 2007). The level of polymorphic loci found in the central Douglas-fir populations are similar to those reported for Picea chihuahuana Mart. (Ledig et al., 1997), Pinus maximartinezii Rzedowski (Ledig et al., 1999), P. pinceana Gordon (Ledig et al., 2001) and P. greggii Engelm. in México (Ramírez et al., 1997; Parraguirre et al., 2002), or Picea breweriana S. Wats. in California and Oregon (Ledig et al., 2005), which also show some extent of geographic isolation, fragmentation and reduced population sizes.
Mean expected heterozygosity (He) per population was relatively low (0.077; Table 2) as compared to genetic diversity values found for Douglas-fir populations in USA and Canada (0.178 in Merkle and Adams, 1987; 0.137 in Li and Adams, 1989). In pine species with reproductive systems similar to that of Douglas-fir, the average He reported is 0.154 (Hamrick and Godt, 1996); thus, genetic diversity for these Douglas-fir populations seems to be rather low, even lower than that found in other Mexican conifers as Picea chihuahuana (Ledig et al., 1997) and Abies religiosa (H.B.K.) Schl. et Cham. (Aguirre et al., 2000). Surprisingly, no significant differences between He and Ho were detected in any Douglas-fir population (Table 2), so there are no indications of heterozygotes deficiency within them, and they appear to be in a mating system equilibrium (see discussion below on the structure of genetic diversity within and among populations).
Northern populations showed He values almost threefold those from central México (Table 2). Jamé and Puerto Palomo in the North had the highest He values (0.175), whereas Tlaxco and San José Capulines in the central region showed the lowest (H < 0.036). Ejido Zapata was completely monomorphic for all loci scored; however, since the sample size was quite low (only 5 mother trees), this might change with a larger number of genotypes.
Genetic structure
Except for locus EST-1, a negative value of F.s was found for all loci, with an average value of -0.068 (Table 3). Thus, under the assumption of random mating, at the sub-population level there is a slight excess of heterozygotes and no inbreeding. In other coniferous species having small and isolated populations it is common to find a dei ciency of heterozygotes, and at least moderate levels of inbreeding, as in the case of P. pinceana (Ledig et al., 2001) and Picea breweriana (Ledig et al., 2005). In that sense, our results are unexpected. Since we used germinated seed only, selection against recessive homozygotes during the early stages of embryo development as proposed in other coniferous species (Desponts and Simon, 1987; Rajora et al., 1998) could account for the heterozygous excess. Mápula-Larreta et al. (2007) found a high proportion of empty seeds in these populations of Douglas-fir, and it was partially attributed to embryo abortion associated to self-pollination in small populations.

The average Fit value was 0.251 (Table 3) indicating that at the whole population level there is a clear deficiency of heterozygous, with a strong trend towards allele fixation at each sub-population. This effect is distinctive of genetic drift acting in small populations as differentiation keeps progressing (Falconer and Mackay, 1996). Average value of the fixation index (Fst) was 0.298 (Table 3), showing that almost one third of the estimated genetic diversity is located among populations. In out-breeding and wind pollinated tree species it is common to find over 85 % of genetic diversity residing within populations (Hamrick and Godt, 1989; Ledig, 1998), so the distribution of genetic diversity in these Douglas-fir populations deviates strongly from this pattern. In fact, the Fst value for these populations is about ive times higher than that estimated for Douglas-fir in other regions (El-Kassaby and Ritland, 1996).
An Fst value above 0.25, as in the present study, indicates that a strong differentiation process among populations is taking place, either from genetic drift or natural selection, in combination with genetic isolation. Similar results have been found in other Mexican conifers with restricted or fragmented natural distribution (Ledig et al., 1997; Aguirre et al., 2000; Parraguirre et al., 2002), so it is no surprising to find this pattern for Mexican Douglas-fir. On average of the 15 loci, the estimated number of migrants per generation ( Nm) was 0.589 (Table 3). Even though it varied up to 7.1 for some loci, the mean value is much lower than that reported for other Douglas-fir populations (El-Kassaby and Ritland, 1996), suggesting that genetic exchange might be strongly limited between populations and genetic drift is likely operating in these small populations.
Geographic pattern of genetic diversity
The average genetic distance between populations was 0.035, but it varied from 0.001 up to 0.125. The highest genetic distances were between populations from the northeast (Jamé or Puerto Palomo) and those from central México. The correlation coefficient between genetic and geographic distances was positive (r = 0.310), and it increased drastically (r = 0.849) when the Mohinora population was excluded from the analysis, leaving only the populations along the Sierra Madre Oriental (Figure 2); however, the Mantel test showed that the association between genetic and geographic distances was not significant in any case. When only the populations from central México were considered, the correlation became negative (r = -0.216); thus, genetic differentiation among these populations seems to be random with respect to their geographic distribution, suggesting again the role of genetic drift in shaping the structure of genetic diversity among them.
Grouping of populations agreed with the geographic trend observed in the correlation analysis (Figure 3a). The two northeastern populations in the Sierra Madre Oriental (Jamé and Puerto Palomo) separated from all others, with high consensus values, whereas those from central México grouped together but with lower consensus values in the subgroups. The Mohinora population joined one of the subgroups from central México with a rather high consensus value; these populations shared similar allele frequencies for most common alleles in 9 of the 15 polymorphic loci surveyed, including three of them fixed (data not shown). The general grouping pattern was not modified when the populations with small sample size were excluded from the analysis, except that the consensus value in the subgroups from central México was higher (Figure 3b).
This grouping pattern is similar to that found using morphological traits in a larger number of populations from the same geographic regions (Reyes-Hernández et al., 2006), except for the Mohinora population. In the Reyes-Hernández et al. (2006) study, as well as in others using shoot phenology traits (Acevedo-Rodríguez et al., 2006), Mohinora was closer to the northeastern populations than to those from central México. The different grouping of the Mohinora population based on morphology and shoot phenology traits as compared with isozyme markers might be due to differences in adaptive value of traits, reflecting the effect of different evolution forces. It is well known that quantitative traits, such as morphology and phenology traits, are commonly subject to natural selection forces whereas isozyme markers are mostly neutral to selection (Lynch et al., 1999; González-Martínez et al., 2002).
CONCLUSIONS
Results of this study show that Mexican Douglas-fir has a high level of genetic diversity and that populations from central México are genetically differentiated from those in the northeast. Since Mexican populations might become an important source of genetic resources for domestication programs of this species, conservation strategies should be delineated to ensure their long-term survival. The marginal distribution of populations in central México, their high degree of differentiation, their highest risk of genetic erosion from fragmentation or climatic change, and their potential value for adaptation to warmer and drier environments, gives them the highest priority for conservation of Douglas-fir within México. Conservation strategies for them, however, should consider the limited production of sound seed and low recruitment capacity of new seedlings, in addition to other biological and environmental pressures that jeopardize in situ conservation in central México.
ACKNOWLEDGEMENTS
The study was supported by the National Council of Science and Technology (CONACYT) of México through projects 33617-B (Genetic diversity and conservation of Pseudotsuga in México) and 2002-COI-6416 of the CONAFOR-CONACYT Fund (Conservation and genetic improvement of Pseudotsuga spp. in Tlaxcala and the central region of México); the study is part of a task undertaken by the Forest Genetic Resources Working Group (FGRWG) of the North American Forestry Commission. We are grateful to the Genetic Marker Lab staff at the Colegio de Postgraduados for helping us to run the samples, particularly to Juan Carlos Zaragoza. We also thank Jean Beaulieu, Natural Resources Canada, for his thorough review and insightful comments to an earlier drat of this paper.
BIBLIOGRAPHY
Acevedo-Rodríguez R, J J Vargas-Hernández, J López-Upton, J Velázquez-Mendoza (2006) Efecto de la procedencia geográfica y de la fertilización en la fenología del brote terminal en plántulas de Pseudotsuga sp. Agrociencia 40:125-137. [ Links ]
Adams W T, D B Neale, A H Doerksen, D B Smith (1990) Inheritance and linkage of isozyme variants from seed and vegetative bud tissues in coastal Douglas-fir (Pseudostuga menziesii var. menziesii (Mirb.) Franco). Silvae Genet. 39:153-167. [ Links ]
Aguirre P E, G R Furnier, L E Eguiarte (2000) Low levels of genetic variation within and high levels of genetic differentiation among populations of species of Abies from southern Mexico and Guatemala. Amer. J. Bot. 87:362-371. [ Links ]
Brown A H D, B S Weir (1983) Measuring genetic variability in plant population: In: Isozymes in Plant Genetics and Breeding. Part "A". S D Tanksley, T J Orton (eds). Elservier, Amsterdan, Holland. pp:219-239. [ Links ]
Brown A H D, C M Hardner (2000) Sampling the gene pools of forest trees for ex situ conservation. In: Forest Conservation Genetics: Principles and Practice. A Young, D Boshier, T Boyle (eds). CSIRO Publishing, Australia. pp:185-196. [ Links ]
Byers J A (1997) Surface distance between two points of latitude and longitude. Computer program available at web site: http://www.wcrl.ars.usda.gov/cec/java/lat-long.htm. [ Links ]
Debreczy Z, I Racz (1995) New species and varieties of conifers from Mexico. Phytologia 78:217-243. [ Links ]
Desponts M, J P Simon (1987) Structure et variabilité génétique de populations d'épinette noire (Picea mariana (Mill.) B. S. P.) dans la zone hémiartique du Nouveau-Québec. Can. J. For. Res. 17:1006-1012. [ Links ]
Domínguez A F A (1994) Análisis histórico-ecológico de los bosques de Pseudotsuga en México. INIFAP-CIR Golfo Centro. Folleto Técnico No. 23. México. 43 p. [ Links ]
El-Kassaby YA, K Ritland (1996) Genetic variation in low elevation Douglas-fir of British Columbia and its relevance to gene conservation. Biodiv. Conserv. 5:779-794. [ Links ]
Falconer D S, T F C Mackay (1996) Introduction to Quantitative Genetics, 4th ed. Longman, New York, USA. 463 p. [ Links ]
Felsenstein J (1995) PHYLIP: a phylogeny inference package, version 3.6. Computer program available at website: http://evolution.genetics.washington.edu/phylip/sotware.html. [ Links ]
Gernandt D S, A Liston (1999) Internal transcribed spacer region evolution in Larix and Pseudotsuga (Pinaceae). Amer. J. Bot. 86:711-723. [ Links ]
González-Martínez S C, R Alia, L Gil (2002) Population genetic structure in a Mediterranean pine (Pinus pinaster Ait.): a comparison of allozyme markers and quantitative traits. Heredity 89:199-206. [ Links ]
Hamrick J L, M J W Godt (1989) Allozyme diversity in plant species. In: Plant Population Genetics, Breeding and Genetic Resources. A H D Brown, M T Clegg, A L Kahler, B S Weir (eds). Sinauer Associates, Sunderland, MA. pp:43-63. [ Links ]
Hamrick J L, M J W Godt (1996) Conservation genetics of endemic plant species. In: Conservation Genetics: Case Histories from Nature. J C Avise, J L Hamrick (eds). Chapman and Hall, New York. pp:281-304. [ Links ]
Hermann R K, D P Lavender (1999) Douglas-Fir planted forests. New For. 17:53-70. [ Links ]
Howe G T, K Jayawickrama, M Cherry, G R Johnson, N C Wheeler (2006) Breeding Douglas-fir. In: Plant Breeding Reviews, Volume 27. J Janick (ed). John Wiley & Sons Inc., New York. pp:245-353. [ Links ]
Jørgensen S, J Hamrick, P V Wells (2002) Regional patterns of genetic diversity in Pinusflexilis (Pinaceae) reveal complex species history. Amer. J. Bot. 89:792-800. [ Links ]
Juárez-Agis A, J López-Upton, J J Vargas-Hernández, C Sáenz-Romero (2006) Variación geográfica en la germinación y crecimiento inicial de plántulas de Pseudotsuga menziesii de México. Agrociencia 40:783-792. [ Links ]
Ledig F T (1998) Genetic variation in Pinus. In: Ecology and Biogeography of Pinus. D M Richardson (ed). Cambridge University Press, Cambridge. pp:251-280. [ Links ]
Ledig F T, V Jacob C, P D Hodgskisss, T Eguiluz P (1997) Evolution and divergence among populations of a rare Mexican endemic, Chihuahua spruce, following Holocene warming. Evolution 51:1815-1827. [ Links ]
Ledig F T, M T Conkle, B Bermejo V, T Eguiluz P, P D Hodgskiss, D R Johnson, W S Dvorak (1999) Evidence for an extreme bottleneck in a rare Mexican pinyon: genetic diversity, disequilibrium, and the mating system in Pinus maximartinezii. Evolution 53:91-99. [ Links ]
Ledig F T, M A Capó A, P D Hodgskiss, H Sbay, C Flores L, M C Thompson, B Bermejo V (2001) Genetic diversity and the mating system of a rare Mexican piñon, Pinuspinceana, and a comparison with Pinus maximartinezii (Pinaceae). Amer. J. Bot. 88:1977-1987. [ Links ]
Ledig F T, P D Hodgskiss, D R Johnson (2005) Genetic diversity, genetic structure, and mating system of brewer spruce (Pinaceae), a relict of the arcto-tertiary forest. Amer. J. Bot. 92:1975-1986. [ Links ]
Li P, W T Adams (1989) Range-wide patterns of allozyme variation in Douglas-fir (Pseudotsuga menziesii). Can. J. For. Res. 19:149-161. [ Links ]
Little E L Jr (1979) Checklist of United States Trees (native and naturalized). USDA Forest Service. Agricultural Handbook No. 541. 375 p. [ Links ]
Lynch M, M Pfrender, K Spitze, N Lehman, J Hicks, D Allen (1999) The quantitative and molecular genetic architecture of a subdivided species. Evolution 53:100-110. [ Links ]
Mápula-Larreta M, J López-Upton, J J Vargas-Hernández, A Hernández-Livera (2007) Reproductive indicators in natural populations of Douglas-fir in Mexico. Biodiv. Conserv. 16:727-742. [ Links ]
Mápula-Larreta M, J López-Upton, J J Vargas-Hernández, A Hernández-Livera (2008) Germinación y vigor de semillas en Pseudotsuga menziesii de México. Ra Ximhai 4:119-134. [ Links ]
Martinez M (1963) Las Pináceas Mexicanas. UNAM. 3ra ed. México, D.F. pp:27-74. [ Links ]
Merkle S A, W T Adams (1987) Patterns of allozyme variation within and among Douglas-fir breeding zones in Southwest Oregon. Can. J. For. Res. 17:402-407. [ Links ]
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590. [ Links ]
Parraguirre L C, J J Vargas H, P Ramírez V, H S Aspiroz R, J Jasso M (2002) Estructura de la diversidad genética en poblaciones naturales de Pinus greggii Engelm. Rev. Fitotec. Mex. 25:279-287. [ Links ]
Rajora O P, L De verno, A Mosseler, D J Innes (1998) Genetic diversity and population of disjunct Newfoundland and central Ontario populations of eastern white pine (Pinus strobus). Can. J. Bot. 76:500-508. [ Links ]
Ramírez H C, J J Vargas H, J Jasso M, G Carrillo C, H Guillén A (1997) Variación isoenzimática en diez poblaciones de Pinus greggii Engelm. Agrociencia 31:223-230. [ Links ]
Reyes-Hernández V J, J J Vargas-Hernández, J López-Upton, H Vaquera-Huerta (2006) Similitud fenotípica de poblaciones mexicanas de Pseudotsuga Carr. Agrociencia 40:545-556. [ Links ]
Slatkin M, N H Barton (1989) A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43:1349-1368. [ Links ]
Stuber C W, J F Wendel, M M Goodman, J S C Smith (1988) Techniques and scoring procedures for starch gel electrophoresis of enzymes from maize (Zea mays L.). North Carolina Agricultural Research Service. North Carolina State University. Raleigh, N.C., U.S.A. Tech. Bull. 286. 87 p. [ Links ]
Swofford D L, R B Selander (1989) BIOSYS-1: a computer program for analysis of allelic variation in population genetics and biochemical systematics, release 1.7. Illinois Natural History Survey, Champaign, Ill. U.S.A. 65 p. [ Links ]
USDA Forest Service (2003) The National Forest Genetics Laboratory (NFGEL) Standard Operating Procedures. Pacific Southwest Research Station. Placerville, CA. 90 p. [ Links ]
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19:395-420 [ Links ]














