Introduction
Xerocomellus Šutara is a genus in the family Boletaceae characterized by boletoid, hypogeous to secotioid basidiomata, usually with small fruit body, sometimes cracked pileus, and smooth to ornamented spores; some species have hypogeous forms with ridged to globose-equinulate spores (Šutara, 2008; Smith et al., 2018; Frank et al., 2020). As other boletes, the species of Xerocomellus form ectomycorrhizas with members of Fagaceae and Pinaceae (Rinaldi et al., 2008; Tedersoo et al., 2010; Wu et al., 2016; Frank et al., 2020). The genus was segregated from Xerocomus Quél. by Šutara (2008), mainly by the basidiospore ornamentation and pileipellis arrangement. Recent phylogenetic studies place Xerocomellus close to Hortiboletus Simonini, Vizzini & Gelardi, in the clade 13 of the Boletoideae (Wu et al., 2016). According to recent research (Frank et al., 2020; Farid et al., 2021) and Index Fungorum (2022) information on the genus, 25 species are recognized worldwide.
Northern Mexico is known for its bolete diversity, with more than 150 species reported from the states of Tamaulipas, Nuevo León, and Coahuila (García-Jiménez and Garza-Ocañas, 2001; García-Jiménez, 2014). So far, no species of Xerocomellus have been described in Mexico using molecular approaches. Only two species are known from this country: Xerocomellus chyrsenteron (Bull.) Šutara and X. truncatus (Singer, Snell & E.A. Dick) Klofac (Saldivar et al., 2021). Due to the diversity of hosts, forest ecosystems and elevations, more fungal species are expected to be found and which need to be described. During mycological exploration in the state of Nuevo León, some noteworthy boletes were found, characterized by the red areolate pileus and boletoid basidiomata. The aim of this study is to describe a new species of Xerocomellus, based on morphological, molecular and ecological data.
Material and Methods
Sampling
The mycological explorations were carried out in the state of Nuevo León, Mexico, during July-August 2009 and July-August 2016 (Fig. 1). The vegetation in the sampling zones is a mixed Pinus-Quercus forest. The identification of the putative host tree was made in the herbarium CFNL, of the Facultad de Ciencias Forestales, Universidad Autónoma de Nuevo León, Mexico. The type of forest was identified according to field observations and satellite images. Methods for collecting, sampling and describing fungi followed Frank et al. (2020).
Hand cut sections were made from dried specimens and mounted in water, KOH 5% and Melzer´s solution for microscopic description. The Methuen Handbook of Colour was used for color terminology (Kornerup and Wanscher, 1978). At least 30 microscopic structures (basidiospores, basidia and pileipellis) were measured with an optical microscope (Motic BA310, San Antonio, USA). The Q ratio, average length (L) and average width (W) were obtained for basidiospores according to Frank et al. (2020). The scanning electron microscope (JEOL JSM-6010PLUS, Tokyo, Japan) of El Colegio de la Frontera Sur (ECOSUR, Chetumal, Mexico) was used to observe basidiospores.
All the specimens are deposited in the mycological collections of the herbaria “José Castillo Tovar” (ITCV) of the Instituto Tecnológico de Ciudad Victoria and CFNL.
Amplification and sequencing
Total DNA was extracted from dried herbarium specimens using a modification of the Murray and Thompson (1980) protocol. The PCR amplification, based on Mullis and Faloona (1987), included 35 cycles with an annealing temperature of 54 ºC, and was carried out with the ITS5 and ITS4 primers (White et al., 1990; Gardes and Bruns, 1993) for the ITS nrDNA region, and the LR0R and LR5 primers (Vilgalys and Hester, 1990; Cubeta et al., 1991) for the 28S nrDNA region (LSU).
The amplicons were verified by agarose gel electrophoresis. The gels were run for 1 h at 95 V cm⁻³ in 1.5% agarose and 1× TAE buffer (Tris Acetate-EDTA). The gel was stained with GelRed (Biotium, USA) and the bands were visualized in an Infinity 3000 transilluminator (Vilber LourmatTM, Marne-la-Vallée, France). The amplified products were purified with the ExoSAP Purification kit (Affymetrix Inc., Santa Clara, USA), following the manufacturer’s instructions. They were quantified and prepared for the sequence reaction using a BigDye Terminator v. 3.1 (Applied Biosystems, Waltham, USA).
These products were sequenced in both directions with an Applied Biosystems model 3730XL (Applied BioSystems, Waltham, USA), at the Instituto de Biología of the Universidad Nacional Autónoma de México (UNAM). The sequences obtained were compared with the original chromatograms to detect and correct possible reading errors.
Phylogenetic analyses
In order to study phylogenetic relationships, our newly produced sequences of three individuals of Xerocomellus were added to reference sequences of ITS and LSU (Table 1) deposited in the NCBI database (GenBank, 2022). Each gene region was independently aligned using the online version of MAFFT v. 7 (Katoh et al., 2002, 2017; Katoh and Standley, 2013). Alignments were reviewed in PhyDE v. 10.0 (Müller et al., 2005), followed by minor manual adjustments to ensure character homology among taxa. The matrices were formed for ITS by 39 taxa (690 characters) and LSU by 23 taxa (592 characters). Hortiboletus was used as outgroup. The aligned matrices were concatenated into a single matrix (39 taxa, 1282 characters) with Mesquite v. 3.2 (Maddison and Maddison, 2021). Phylogenetic inferences were estimated with maximum likelihood in RAxML v. 8.2.10 (Stamatakis, 2014) with a GTR + G model of nucleotide substitution. To assess branch support, 1000 rapid bootstrap replicates were run with the GTRCAT model. For Bayesian posterior probability, the best evolutionary model for alignment was sought using PartitionFinder v. 2 (Lanfear et al., 2014, 2017; Frandsen et al., 2015).
Table 1: GenBank accession numbers corresponding to the sequences used in the phylogenetic analyses. In bold the accessions of the new species generated for this study.
| Species name | Isolate/Voucher/strain | Locality | GenBank Accessions | |
|---|---|---|---|---|
| ITS | nrLSU | |||
| Xerocomellus aff. chrysenteron (Bull.) Šutara | JLF5684 | USA | MH168533 | ----- |
| Xerocomellus amylosporus (A.H. Sm.) J.L. Frank & N. Siegel | JLF3012 | USA | KM213635 | KU144742 |
| Xerocomellus atropurpureus J.L. Frank, N. Siegel & C.F. Schwar | JLF3620 | USA | KU144749 | KU144750 |
| Xerocomellus atropurpureus J.L. Frank, N. Siegel & C.F. Schwar | JLF4664 | USA | KY659589 | ----- |
| Xerocomellus atropurpureus J.L. Frank, N. Siegel & C.F. Schwar | NS120712 | USA | KM213641 | KM213642 |
| Xerocomellus atropurpureus J.L. Frank, N. Siegel & C.F. Schwar | JLF2795 | USA | KM213638 | KM213639 |
| Xerocomellus behrii (Harkn.) Castellano, M.E. Sm. & J.L. Frank | OSC Trappel 17620 | ----- | KJ882290 | ----- |
| Xerocomellus bolinii J.A. Bolin, A.E. Bessette, A.R. Bessette, L.V. Kudzma, J.L. Frank & A. Farid | JAB 133 | USA | MW675729 | MW662582 |
| Xerocomellus bolinii J.A. Bolin, A.E. Bessette, A.R. Bessette, L.V. Kudzma, J.L. Frank & A. Farid | JAB 95 | USA | MW675735 | MW662589 |
| Xerocomellus bolinii J.A. Bolin, A.E. Bessette, A.R. Bessette, L.V. Kudzma, J.L. Frank & A. Farid | JAB 110 | USA | MW675735 | MW662580 |
| Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente | 18219 Type ITCV | Mexico | ON392096 | ON254917 |
| Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente | 5193 ITCV | Mexico | ON392097 | ON254918 |
| Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente | 5192 ITCV | Mexico | ON392098 | ON254919 |
| Xerocomellus cisalpinus (Simonini, H. Ladurner & Peintner) Klofac | KR-M-0044831 | Germany | MT006036 | ----- |
| Xerocomellus cisalpinus (Simonini, H. Ladurner & Peintner) Klofac | LUGO:ECC19102906 | Spain | MW376718 | ----- |
| Xerocomellus diffractus (Simonini, H. Ladurner & Peintner) Klofac | JLF3554 | USA | KU144769 | KU144770 |
| Xerocomellus diffractus (Simonini, H. Ladurner & Peintner) Klofac | JLF5745 | USA | MH168534 | ----- |
| Xerocomellus diffractus (Simonini, H. Ladurner & Peintner) Klofac | NS120612 | USA | KM213650 | KM213651 |
| Xerocomellus dryophilus (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | CFS3Nov11-2 | USA | KM213645 | KX534074 |
| Xerocomellus dryophilus (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | JLF4134 | USA | KX534076 | KY659593 |
| Xerocomellus mcmurphyi (Zeller & C.W. Dodge) Castellano, Saylor, M.E. Sm. & J.L. Frank | OSCMES 282 | ----- | KJ882289 | KJ882292 |
| Xerocomellus mendocinensis (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | JLF2275 | USA | KM213653 | KM213654 |
| Xerocomellus mendocinensis (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | JLF3558 | USA | KU144785 | KU144786 |
| Xerocomellus mendocinensis (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | CFS 1Nov11 | USA | KM213656 | KM213657 |
| Xerocomellus mendocinensis (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank | HDT 18392 | USA | KM213655 | ----- |
| Xerocomellus poederi G. Moreno, Heykoop, Esteve-Rav., P. Alvarado & Traba | AH 44050 Type | ----- | NR155971 | ----- |
| Xerocomellus pruinatus (Fr. & Hök) Šutara | G.M. 2015-09-23.4 | Luxembourg | MW603181 | MW603181 |
| Xerocomellus rainisie (Bessette & O.K. Mill.) N. Siegel, C.F. Schwarz & J.L. Frank | OKM25915 | USA | KM213664 | ----- |
| Xerocomellus ripariellus (Redeuilh) Šutara | 301 | Spain | MN685108 | ----- |
| Xerocomellus salicicola C.F. Schwarz, N. Siegel & J.L. Frank | CS-5Mar2014-1 | ----- | KU144791 | KU144792 |
| Xerocomellus salicicola C.F. Schwarz, N. Siegel & J.L. Frank | UCSC1028 | ----- | KU144793 | KU144794 |
| Xerocomellus sarnarii Simonini, Vizzini & U. Eberh. | ML900101XE | Cyprus | MH011930 | MH011930 |
| Xerocomus truncatus (Singer, Snell & E.A. Dick) Klofac | HDT22426 | USA | KU144796 | ----- |
| Xerocomus truncatus (Singer, Snell & E.A. Dick) Klofac | NY13857 | USA | KU144795 | ----- |
| Xerocomellus zelleri (Murrill) Klofac | JLF2977 | USA | KM213666 | ----- |
| Xerocomellus zelleri (Murrill) Klofac | Murrill Type | USA | KU144803 | ----- |
| Hortiboletus campestris (A.H. Sm. & Thiers) Biketova & Wasser | DD614 | USA | MH168538 | ----- |
| Hortiboletus cf. rubellus (Krombh.) Simonini, Vizzini & Gelardi | JLF3093 | USA | KU144805 | ----- |
Phylogenetic analyses were performed using MrBayes v. 3.2.6 x64 (Huelsenbeck and Ronquist, 2001). The information block for the matrix included two simultaneous runs, four Montecarlo chains, temperature set to 0.2 and sampling 10 million generations (standard deviation ≤0.1) with trees sampled every 1000 generations. The first 25% of samples were discarded as burn-in, and stationarity was checked in Tracer v. 1.6 (Rambaut et al., 2014). Trees were visualized and optimized in FigTree v. 1.4.4 (Rambaut, 2014). The phylogenetic tree is available at TreeBASE (accession number: TB2:S16457).
Results
Molecular analyses
The two simultaneous Bayesian runs continued until the convergence parameters were met, and the standard deviation fell below 0.001 after 4.5 million generations (Fig. 2). No significant changes in tree topology trace or cumulative split frequencies of selected nodes were observed after about 0.25 million generations, which were discarded as 25% burn-in. The analysis produced a phylogenetic tree where the new species is shown as a monophyletic clade with strong statistical support (1 Bayesian Posterior Probability (PP) and 100% bootstrap proportion (BP) for Maximum Likelihood).
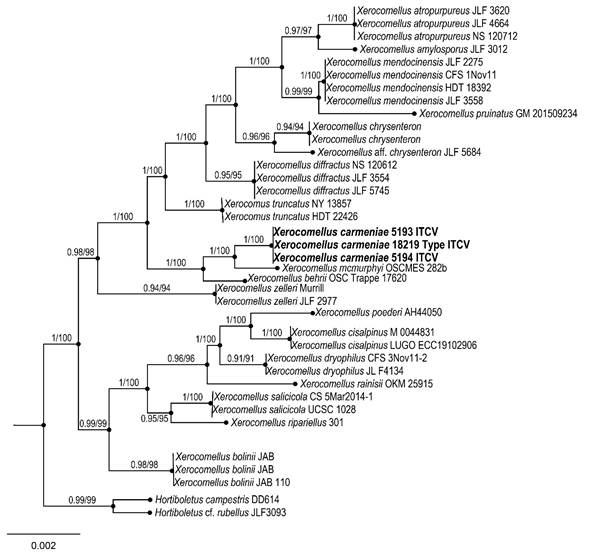
Figure 2: Bayesian inference phylogram of ITS-LSU sequence data. Posterior probability (left of slash) from Bayesian analysis and Bootstrap support
Taxonomy
Basidiomycota
Agaricomycetes
Boletales
Boletaceae
Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente sp. nov. Figs. 3, 4.

Figure 3: Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente (holotype). A-B. details of the basidiomata; C. details of the tubes; D. context; E. basidiomata and details of the pileus. Scale: 10 mm.

Figure 4: Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente (holotype) A-B. basidiospores; C-D. elements of pileipellis. Scale=A:10 µm; B: 5 µm; C-D: 10 µm.
TYPE: MEXICO. Nuevo León, municipality Iturbide, Puerto Pastores, Pinus-Quercus forest, under Q. canbyi, 16.VII.2016, J. García 18219 (holotype: ITCV!, isotype: CFNL!). Mycobank: MB843147. Genbank: ITS: ON392096; LSU: ON254917.
Xerocomellus carmeniae is characterized by the following combination of features: boletoid basidioma, reddish areolate pileus, context yellowish, staining blue when cut, truncate, elongate to ellipsoid basidiospores, 10.5-13.6 × 5.7-7.8 µm.
sGene sequences ex-holotype. MF616521 (tef-1α), MF616525 (nLSU).
Basidiomata boletoid, sometimes with a poorly developed stipe; pileus 18-32 mm diameter, vivid red to deep red (11A8-11C8), light brown to brown (6D4-6E4) background pale yellow (4A6), slightly convex to flattened, margin irregular; context yellowish (4A8), bruising greyish green (28B5) to greyish blue (24D5) when young, unchanging when mature; tubes irregularly arranged, emarginated sometimes, yellowish (30A8) to greyish green (27E5), with angular to irregular pores, sometimes covered by a thin whitish layer near the stipe apex; stipe 13-34 × 0.5-10 mm, irregular and sinuous, pale brown (5D4) to pale yellow (4A7), with small reddish (11A8) scales, sometimes becoming pale brown (5D4) to grey brown (5D3) at the slender base; pileipellis 105-174 µm thick, composed of compacted interwoven chains of cylindrical to subglobose hyphae, 10-22 µm diameter, with erect and clavate to fusoid terminal cells, brownish to yellowish in KOH 5% and Melzer’s reagent, with granular intracellular content; hymenophoral trama slightly divergent, with cylindric and loosely interwoven hyphae, 2-6 µm in diameter, hyaline, thin-walled; basidia clavate to subclavate, 25-36 × 11-18 µm, hyaline, 4-spored, with sterigma projecting up to 3 µm long, thin-walled; hymenial cystidia not observed; basidiospores 10.5-13.6 × 5.7-7.8 µm (L=11.05, W=6.90, Q=1.60, N=30), truncate, elongate, brown in KOH and Meltzer, guttulate, sometimes truncate, smooth and thick-walled.
Habit and habitat: scattered in small groups, mixed Pinus-Quercus forest under Quercus canbyi Trel.
Distribution: only known from Nuevo León, Mexico.
Etymology: named carmeniae in honor of María del Carmen A. Medina Cortés, who found the specimens of this study.
Additional material examined: MEXICO. Nuevo León, municipality Iturbide, Puerto Pastores, 02.VIII.2009, F. Garza-Ocañas 5192 (CFNL), 5193 (CFNL).
Notes: the new species differs from other members of Xerocomellus such as Xerocomellus macmurphyi (Zeller & C.W. Dodge) Castellano, Saylor, M.E. Sm. & J.L. Frank and Xerocomellus behrii (Harkn.) Castellano, M.E. Sm. & J.L. Frank, due to the globose and hypogeous basidiomata with spinose basidiospores (Smith et al., 2018; Frank et al., 2020). It also differs from the boletoid to secotioid basidiomata of X. amylosporus (A.H. Sm.) J.L. Frank & N. Siegel, because it has amyloid and larger basidiospores (Frank et al., 2020).
Discussion
Boletaceae is one of the most common groups of fungi present in temperate forest (Ayala-Vásquez et al., 2022). Despite the usually developed boletoid, lamellate, epigeous fruit body, several species grow underground as false truffles or secotioid species (Thiers and Trappe, 1969; Binder and Hibbett, 2006; Smith et al., 2018). Most of them form ectomycorrhizal associations with angiosperms and gymnosperms (Hasselquist et al., 2011; Wu et al., 2016). The boletes are one of the mayor groups of fungi present in northeastern Mexican temperate forest with about 150 described species (García-Jiménez, 2014). More species are being described from the Mexican temperate zone, indicating the great species richness in Mexico (Ayala-Vásquez et al., 2018).
The genus Xerocomellus shows typically epigeous boletoid taxa; nevertheless, few hypogeous and secotioid species are known (Smith et al., 2018; Frank et al., 2020). The most remarkable features of the new species are the boletoid basidiomata, the reddish pileus, and the truncate basidiospores. Interestingly, Xerocomellus carmeniae is phylogenetically closer to hypogeous species such as Xerocomellus macmurphyi and Xerocomellus behrii, than to secotioid species such as Xerocomellus amylosporus or with truncate spores such as Xerocomellus mendocinensis (Thiers) N. Siegel, C.F. Schwarz & J.L. Frank (Frank et al., 2020). Despite Xerocomellus carmeniae has a boletoid basidiomata, some collections show an aberrant form that could represent a transition to a secotioid form (Fig. 5). Even when the hymenium is not completely enclosed within a persistent peridium, the absence of hymenial cystidia and the basidiospores without hylar depression could indicate a possible loss of the capability for active dispersal. These characteristics are found in the truly secotioid boletes (Thiers and Trappe, 1969; Thiers, 1984). Other Xerocomellus species such as X. amylosporus and X. atropurpureus J. L. Frank, N. Siegel & C.F. Schwarz also have a gastroid morphological variation (Frank et al., 2020). More collections are needed to clarify the possible secotioid transition of this new species.

Figure 5: Xerocomellus carmeniae Garza-Ocañas, J. García & de la Fuente (F. Garza-Ocañas 5192) showing an aberrant basidioma. Scale=10 mm.
The sequestrate Boletales are still poorly studied when compared with their epigeous relatives. In Mexico, only some species of Octaviania Vittad., Melanogaster Corda and Rhizopogon Fr. (Trappe and Guzmán, 1971; Cázares et al., 1992; 2008) have been described. Considering Mexico as a diversification center of the genera Quercus L. and Pinus L, trees which form association with several species of mycorrhizal fungi (Trappe et al., 2009), more studies about the Mexican symbiotic fungi are recommended.











 nueva página del texto (beta)
nueva página del texto (beta)




