Introduction
Lysiloma Benth. (Bentham, 1844) is one of 36 genera within the tribe Ingeae (Brown, 2008) in the extended subfamily Caesalpinioideae (LPWG, 2017). Two recent phylogenetic analyses (Brown et al., 2008; Iganci et al., 2015) suggest that Lysiloma and the monotypic Hesperalbizia Barneby & J.W. Grimes are sister taxa. As currently understood, it is composed of eight tree species with a primarily Neotropical distribution, ranging from Arizona and Florida through Mexico, Central America, and the Antilles (Thompson, 1980; Gale and Pennington, 2004; Andrade and Sousa, 2012) (Table 1).
Table 1: General distribution of the genus Lysiloma Benth. We included L. divaricatum (Jacq.) Macbr. and L. microphyllum Benth. as two species following Thompson’s manuscript (1980), but Gale and Pennington (2004) considered them coespecific. Our results support Gale and Pennington’s point of view.
| Species (according to Thompson, 1980) | Distribution and elevation in m above sea level |
| Lysiloma acapulcense (Kunth) Benth. | Mexico, Central America to El Salvador, and Honduras. 600-2000 |
| Lysiloma auritum (Schltdl.) Benth. | Southwest Mexico to Central America except Belize. 100-2400 |
| Lysiloma candidum Brandegee | Mexico (Baja California Norte, Baja California Sur, and Sonora). 0-400 |
| Lysiloma divaricatum (Jacq.) Macbr. | Mexico (Chiapas, Oaxaca, and Veracruz), Central America to Costa Rica. 20-1700 |
| Lysiloma latisiquum (L.) Benth. | USA (Florida), southeast Mexico, Belize, Guatemala, and the Bahamas, Cuba, and Hispaniola. 0-400 |
| Lysiloma microphyllum Benth. | Mexico (Baja California, Chihuahua, Colima, México, Morelos, Nayarit, Oaxaca, Querétaro, San Luis Potosí, Sonora, and Tamaulipas). 0-1600 |
| Lysiloma sabicu Benth. | The Bahamas, Cuba, and Hispaniola. 50-900 |
| Lysiloma tergeminum Benth. | Mexico (Colima, Guerrero, México, Michoacán, Morelos, Nayarit, and Puebla). 0-2000 |
| Lysiloma watsonii Rose | USA (Arizona) and Mexico (Chihuahua, and Sinaloa). 300-1300 |
Lysiloma acapulcense (Kunth) Benth. and L. divaricatum (Jacq.) J.F. Macbr. are the species with the widest distribution, ranging from northern Mexico to Central America (Nicaragua and Costa Rica). Both taxa include a long list of synonyms and broad morphological variation. These taxa urgently require a deeper future reassessment. The rest of the species have more restricted distributions (Table 1). The general distribution of the genus coincides in general terms with Megamexico 3, a biogeographic area that includes the entire Mexican territory, but also the southern part of the United States of America (the northern portions of the Chihuahuan and Sonoran deserts in California, Arizona, New Mexico, and Texas) in the north, and Central American to Nicaragua north of the lakes, in the south (Rzedowski, 1991). Two exceptions are L. latisiliquum (L.) Benth., and L. sabicu Benth., which are found in Florida, the Bahamas, and the West Indies (Thompson, 1980). In general, Lysiloma species occupy a wide variety of dry forest types, being dominant or codominant elements in many of them (Rascón-Ayala et al. 2018; Ancona et al., 2019; Ortiz-Ávila et al., 2020).
The generic name of this group of plants derives from the Greek Lysis (to lose) and loma (edge), referring to the shedding of the legume edge at fruit maturity that allows for the dispersion of the seeds. This type of dehiscence has been referred to as craspedial (Thompson, 1980; Gale and Pennington, 2004). Two distinctive species of the genus lack this feature, Lysiloma sabicu and L. latisiliquum (Fig. 1E, F), where the valves remain together at maturity. An additional, distinctive feature of the genus is the membranous, foliaceous stipule (Fig. 1C) (Bentham, 1875), a character state that is shared by Hesperalbizia. Thompson (1980) proposed an informal classification system with two subgenera based upon fruit dehiscence characters: Lysiloma subg. Lysiloma Thompson (nom. nud.), with indehiscent or late dehiscent legumes and Lysiloma subg. Lysivalva Thompson (nom. nud.), where the legume is dehiscent, and the valves break apart at the margins. Subgenus Lysiloma is composed of two sections defined by inflorescence types (capitulum vs. raceme) and flower pedicels (sessile vs. pedicellate): Lysiloma subg. Lysivalva sect. Capitata Thompson (nom. nud.), and sect. Racemosa Thompson (nom. nud.) (Fig. 1A, B, D).

Figure 1: General morphology of Lysiloma Benth. A. leaves, and inflorescences of Lysiloma acapulcense (Kunth) Benth.; B. inflorescence of L. acapulcense; C. leave and stipule of Lysiloma latisiliquum (L.) Benth.; D. inflorescence of Lysiloma latisiliquum; E. young and old fruits of L. latisiliquum; F. fruit of Lysiloma sabicu Benth. Pictures A, B: Claudia Ramírez; C, D, E: Rodrigo Duno de Stefano; F: Susan Ford Collins.
As is the case of most members of tribe Ingeae, Lysiloma pollen grains are arranged in polyads of 16 (28-32 in L. divaricatum (Jacq.) J.F. Macbr.) (Guinet and Grimes, 1997). In general, polyads are 46-79 µm diameter, isodiametric where the eight radial pollen grains are longer than wide, whereas the internal tetrad is composed of square pollen grains. The external pollen grains are dissymmetric in thickness; whereas the tectum is perforated with 20-100/µm2 ornamentation is uniform throughout, fossulate, or polygonal with more or less rounded areoles (Sorsa, 1969; Guinet and Grimes, 1997). Hesperalbizia occidentalis (Brandegee) Barneby & J.W. Grimes is characterized by similar yet larger, symmetrical polyads. Both Lysiloma and Hesperalbizia feature 2n=26 chromosomes (Thompson, 1980; Rico Arce, 1992).
Lysiloma is known from a Tertiary (Olygocene) fossil record, L. mixtecanaMagallón-Puebla & Cevallos-Ferriz (Magallón-Puebla and Cevallos-Ferriz, 1993). This fossil was recovered in a sedimentary layer in Puebla, along with records of other members of tribe Ingeae: Pithecellobium grimesii Calvillo-Canadell & Cevallos-Ferriz and Pithecellobium barneby Calvillo-Canadell & Cevallos-Ferriz (Calvillo-Canadell and Cevallos-Ferriz, 2005).
Assessing the monophyly of Lysiloma is particularly relevant since two related genera studied from a molecular perspective were non-monophyletic: Abarema Pittier (Iganci et al., 2015), and Zygia P. Brown (Ferm et al., 2019). Aiming at testing the monophyly and sister-group relationships of Lysiloma, we performed phylogenetic analyses combining morphological and molecular datasets. We employed morphological characters, plastid (trnK and matK), and nuclear (ETS) DNA sequence data.
These analyses were carried out with parsimony for the morphological characters and parsimony and Bayesian inference for the molecular and total evidence approach. This study will allow us to test three hypotheses: 1) Lysiloma is monophyletic as currently circumscribed; 2) Lysiloma and Hesperalbizia are closely related (Brown et al., 2008; Iganci et al., 2015), and 3) the infrageneric relationships between the species as proposed by Thompson (1980). Regarding infrageneric relationships, the informal classification proposal of Thompson (1980) was used as a null hypothesis. Moreover, we were interested in estimating the chronology of the diversification under the uncorrelated lognormal relaxed molecular clock approach. To do so, we adopt the same assumption of Becerra (2005) regarding Bursera Jacq. ex L. (Burseraceae) and the expansion of the dry forest. In other words, the history and evolution of Bursera in Mexico mirrors the history of the dry forest in Mexico. Thus, the very forces that drove the range expansions and contractions of the dry forests also shaped the diversification of Bursera. Because Lysiloma is highly adapted to the ecological conditions of the dry forest, we test whether the diversification of this genus is related to the expansion of the dry forest in the Miocene (20-5 Mya (million years ago)), when arid environments expanded across the world.
Material and Methods
Taxon sampling
The ingroup includes the nine Lysiloma species recognized by Thompson (1980) and Hesperalbizia occidentalis whereas the outgroup is composed of basal members of the Mimosoid clade, including Acaciella angustissima (Mill.) Britton & Rose, Calliandra eriophylla Benth., C. haematocephala Hassk., Faidherbia albida (Delile) A. Chev., Mariosousa dolichostachya (S.F. Blake) Seigler & Ebinger, Senegalia parviceps (Speg.) Seigler & Ebinger, Vachellia farnesiana (L.) Wight & Arn., Zapoteca formosa (Kunth) H.M. Hern., and Zapoteca tetragona (Willd.) H.M. Hern. (Table 2). An attempt was made to include two or three accessions per species to sample their geographical and ecological range. However, some species feature wide distributions (e.g. Lysiloma acapulcense) and it is possible that an even broader sampling may be required to represent the morphological and distributional range of these particular species.
Table 2: Summary of the botanical material used for molecular analysis of the genus Lysiloma Benth., including accessions extracted from GenBank. In bold GenBank accession numbers of sequences generated in the present study.
| Species | ETS | matK | trnK |
|---|---|---|---|
| Acaciella angustissima (Mill.) Britton & Rose | EF638082.1 | HM020733.1 | E. López 1128 (CICY) MN755794 |
| Calliandra eriophylla Benth. | E. López 1099 (CICY) MN755770 | EU025883.1 | E. López 1099 (CICY) MN755797 |
| Calliandra haematocephala Hassk. | Bot. Garden Fairchild 2007 0163A MN755769 | MH749029.1 | R. Duno 2425 (CICY) MN755796 |
| Faidherbia albida (Delile) A. Chev. | EF638163.1 | JF270778.1 | AF274129 |
| Hesperalbizia occidentalis (Brandegee) Barneby & J.W. Grimes | R. García Sosa 71 (MO) MN755772 | R. García Sosa 71 (MO) MN755817 | R. García Sosa 71 (MO) MN755799 |
| Hesperalbizia occidentalis (Brandegee) Barneby & J.W. Grimes | Santana and Cervantes 868 (ZEA) MN755773 MN755774 | A. Sánchez and A. Nava 399 (ZEA) MN755818 MN755819 | C. E. Hughes 1543 (MEXU) MN755800 MN755801 |
| Lysiloma acapulcense (Kunth) Benth. | J. Calónico Soto 7192 (FCME) MN755778 MN755779 | O. Alcántara and M. Paniagua 5831 (FCME) MN755822 | O. Alcántara and M. Paniagua 5831 (FCME) MN755803 |
| Lysiloma acapulcense (Kunth) Benth. | - | E. López s.n (CICY) MN755823 | E. López s.n (CICY) MN755804 |
| Lysiloma auritum (Schltdl.) Benth. | Bot. Garden Fairchild 62265 MN755780 MN755781 | Bot. Garden Fairchild 62265 MN755824 | Bot. Garden Fairchild 62265 MN755805 |
| Lysiloma auritum (Schltdl.) Benth. | - | JQ587745.1 | - |
| Lysiloma candidum Brandegee | - | KX302335.1 | - |
| Lysiloma divaricatum (Jacq.) J.F. Macbr. | M. Ayala et al. 918 (FCME) MN755783 MN755784 | M. Ayala et al. 918 (FCME) MN755826 | M. Ayala et al. 918 (FCME) MN755807 |
| Lysiloma divaricatum (Jacq.) J.F. Macbr. | - | - | AF523088.1 |
| Lysiloma latisiliquum (L.) Benth. | P. Simá 2287 (CICY) MN755785 MN755786 MN755787 | P. Simá 2287 (CICY) MN755827 MN755828 | P. Simá 2287 (CICY) MN755808 |
| Lysiloma latisiliquum (L.) Benth. | - | - | S. Villanueva s.n. (CICY) MN755809 MN755810 |
| Lysiloma microphyllum Benth. | J. Calónico Soto 9422 (FCME) MN755788 MN755789 | I. Rosas et al. 2284 (MO) MN755829 | I. Rosas et al. 2284 (MO) MN755811 |
| Lysiloma microphyllum Benth. | - | M. Ordóñez 2 (CHIP) MN755830 | - |
| Lysiloma microphyllum Benth. | E. López 1134 (CICY) MN755790 | E. López 1134 (CICY) MN755831 | E. López 1134 (CICY) MN755812 MN755813 |
| Lysiloma sabicu Benth. | Bot. Garden Fairdchild 2012-039 MN755775 | Bot. Garden Fairdchild 2012-039 MN755820 MN755821 | Bot. Garden Fairdchild 2012-039 MN755802 |
| Lysiloma sabicu Benth. | J. R. Abbott 24059 (MO) MN755776 MN755777 | - | |
| Lysiloma tergeminum Benth. | S. Valencia 4058 (FCME) MN755791 MN755792 | - | S. Valencia 4058 (FCME) MN755814 |
| Lysiloma tergeminum Benth. | - | - | EU812062.1 |
| Lysiloma tergeminum Benth. | J. Calónico Soto 84 (FCME) MN755793 | J. Calónico Soto 84 (FCME) MN755832 MN755833 MN755834 | J. Calónico Soto 84 (FCME) MN755815 |
| Lysiloma watsonii Rose | Regional Bot. Garden Roger Orellana (RD-001) MN755782 | Regional Bot. Garden Roger Orellana (RD-001) MN755825 | Regional Bot. Garden Roger Orellana (RD-001) MN755806 |
| Mariosousa dolichostachya (S.F. Blake) Seigler & Ebinger | EF638084.1 | EU812056.1 | AF523190.1 |
| Senegalia parviceps (Speg.) Seigler & Ebinger | L. E. Quispe 63 (MO) MN755767 MN755768 | L. E. Quispe 63 (MO) MN755816 | L. E. Quispe 63 (MO) MN755795 |
| Vachellia farnesiana (L.) Wight & Arn. | EF638128.1 | FJ711552.1 | AY574103.1 |
| Zapoteca formosa (Kunth) H.M. Hern. | J. Peñaranda 216 (MO) MN755771 | AY125854.1 | R. Duno s.n. (CICY) MN755798 |
| Zapoteca tetragona (Willd.) H.M. Hern. | EF638133.1 | JQ587912.1 | AF523097 |
DNA extraction, amplifications, and sequencing
For the molecular analyses, fresh leaflet tissue collected in the field, in the Regional Botanical Garden Roger Orellana of the Centro de Investigación Científica de Yucatán, A. C., and herbarium material were employed. Herbarium specimens used in these analyses came from CICY, FCME, MA, MEXU, MO, UCOL, and ZEA (acronyms as in Thiers, 2020 continuously updated). Permission was requested from herbarium curators to use leaflet material for DNA extraction. Sixty-five new sequences were generated (17 ETS: MN755767-MN755793, 19 trnK: MN755794-MN755815, and 14 matK: MN755816-MN755834). Additional sequences were downloaded from GenBank (GenBank, 2020), particularly from datasets created by Miller and Bayer (2001), Miller et al. (2003), Brown et al. (2008), and Heil et al. (2009). For GenBank accessions see Supplementary Material (Table 2, 3).
Table 3: Additional accessions for the large ETS analysis.
| Taxon | GenBank’s accession number |
|---|---|
| Abarema jupunba (Willd.) Britton & Killip | EF638109, EF638110 |
| Abarema piresii Barneby & J.W. Grimes | KF921624 |
| Acacia chartacea Maslin | DQ029304, DQ029305 |
| Acacia dempsteri F. Muell. | DQ029300 |
| Acacia karina Maslin & Buscumb | KC796100 |
| Acacia pyrifolia DC. | DQ029293 |
| Acacia ryaniana Maslin | DQ029303 |
| Acacia semicircinalis Maiden & Blakely | KC283889 |
| Acacia strongylophylla F. Muell. | DQ029299 |
| Acacia victoriae Benth. | DQ029310, DQ029311 |
| Acaciella angustissima (Mill.) Britton & Rose | EF638082.1 |
| Albizia adinocephala (Donn. Sm.) Britton & Rose ex Record | EF638144 |
| Albizia polycephala (Benth.) Killip, | KF921625 |
| Archidendron ellipticum (Blume) I.C. Nielsen | EF638153 |
| Archidendron hendersonii I.C. Nielsen | HM800427 |
| Archidendron kanisii R.S. Cowan | EF638098 |
| Archidendron lucyi F. Muell. | HM800428 |
| Archidendron whitei I.C. Nielsen | EF638099 |
| Blanchetiodendron blanchetii (Benth.) Barneby & J.W. Grimes | KF921626 |
| Cojoba arborea (L.) Britton & Rose | EF638108, EF638095 |
| Cojoba undulatomarginata L. Rico | EF638096 |
| Ebenopsis confinis (Standl.) Britton & Rose | KF921650, EF638100 |
| Ebenopsis ebano (Berland.) Barneby & J.W. Grimes | EF638101, EF638102, KF921651 |
| Enterolobium contortisiliquum (Vell.) Morong | EF638151 |
| Enterolobium gummiferum (Mart.) J.F. Macbr. | KF921652 |
| Enterolobium timbouva Benth. | KF921654 |
| Faidherbia albida (Delile) A. Chev. | EF638163.1 |
| Falcataria moluccana (Miq.) Barneby & J.W. Grimes | HM800429, HM800430 |
| Havardia mexicana (Rose) Britton & Rose | KF921655 |
| Havardia pallens (Benth.) Britton & Rose | EF638146, EF638147, KF921656 |
| Hydrochorea corymbosa (Rich.) Barneby & J.W. Grimes | KF921657 |
| Inga thibaudiana DC. | KF921659 |
| Leucochloron bolivianum C.E. Hughes & Atahuachi | KF921660 |
| Macrosamanea pubiramea (Steud.) Barneby & J.W. Grimes | KF921665 |
| Mariosousa dolichostachya (S.F. Blake) Seigler & Ebinger | EF638084.1 |
| Pararchidendron pruinosum (Benth.) I.C. Nielsen | EF638129 |
| Paraserianthes lophantha (Willd.) I.C. Nielsen | KU727929, KU727943, HM800432 |
| Paraserianthes toona (Bailey) I.C. Nielsen | EF638106, EF638107 |
| Pithecellobium diversifolium Benth. | KF921666 |
| Pithecellobium dulce (Roxb.) Benth. | EF638142, EF638143 |
| Samanea saman (Jacq.) Merr. | KF921668 |
| Samanea tubulosa (Benth.) Barneby & J.W. Grimes | EF638135 |
| Sphinga acatlensis (Benth.) Barneby & J.W. Grimes | KF921669, EF638145 |
| Thailentadopsis nitida (Vahl) G.P. Lewis & Schrire | KF921670 |
| Vachellia farnesiana (L.) Wight & Arn. | EF638128.1 |
| Wallaceodendron celebicum Koord. | EF638097 |
| Zapoteca tetragona (Willd.) H.M. Hern. | EF638133.1 |
| Zygia racemosa (Ducke) Barneby & J.W. Grimes | KF921671 |
The matK gene has been among the most useful loci for resolving plant phylogenetic relationships at different evolutionary timescales (Hilu et al., 2008). It has been used to assess and monitor biodiversity and, via community phylogenetics, to investigate ecological and evolutionary processes that may be responsible for the community structure of particular forests (DNA barcoding) (Heckenhauer et al. 2017). The trnK intron sequences also provide similar levels of phylogenetic information as matK. Combining the trnK with matK increases overall bootstrap support (Hilu et al., 2008). The external transcribed spacer (ETS) of 18S-26S nuclear ribosomal DNA has been used intensely in phylogenetic studies of the tribes Acacieae and Ingeae, generally together with ITS (Brown et al., 2008; Murphy et al., 2010). The ETS has also a high rate of sequence evolution by nucleotide substitution (Baldwin and Markos, 1998).
Total DNA from leaflets (fresh or from herbarium material) was obtained with the DNeasy Plant Mini Kit (QIAGEN Inc., Valencia, California) following the provider’s specifications. To assess concentration and relative quality of DNA, 3 µl of the final volume plus 2 µl loading buffer were run for 30 minutes at 6 volt/cm in a 1% agarose gel prepared with TBE 0.5X. The resulting gel was revealed by immersion for 20-30 minutes in a 0.1 µg/ml ethidium bromide solution and later observed in a DigiDoc-It Imaging System (v. 6.7.1; UVP, Inc., Cambridge, UK) transilluminator. Furthermore, DNA purity and concentration were quantified with a NanoDrop 2000c (Thermo Scientific™, Waltham, USA). Then, DNA samples were standardized at 10 ng µl-1. Amplifications were performed in an Applied Biosytems Veriti 96 Well Thermal Cycler (Applied Biosystems, Foster City, USA). Volumes of reagents and conditions for the amplifications were as follows:
ETS: 30 µl of mix containing 3 µl 10X Buffer, 2.5 µl MgCl2, 0.6 µl (~10 ng) primer, 4 µl Q solution, 1 µl 1.25 mM l-1 dNTP, 0.2 µl (1 U) TAQ polymerase, 2 µl (~10 ng) DNA, then completed to volume (approx. 16.1 µl) with ultra-pure water. PCR’s were conducted under the following protocol: 94 °C for 3 min + 30 cycles (94 °C for 1 min + 60.5 °C for 1 min + 72 °C for 2 min) + 72 °C for 7 min. Primers were 18S-IGS and 26S-IGS (Baldwin and Markos, 1998).
trnK: 20 µl containing 2.0 µl 10X Buffer, 0.8 µl of MgCl2, 1 µl (~ 10 ng) of primers, 0.8 µl MgCl2, 1 µl (~ 10 ng) primers, 4 µl Q solution, 1.5 µl 1.25 mM l-1 of dNTP, 0.2 µl (1 U) TAQ polymerase, 3 µl (~10 ng) DNA, then completed to volume (approx. 6.5 µl) with ultra-pure water. PCR’s were conducted under the following protocol: 94 °C for 3 min + 30 cycles of (94 °C for 1 min + 55 °C for 1 min + 72 °C for 2 min) + 72 °C for 7 min. Primers were trnK 3914 and Ac 283R (Johnson and Soltis, 1994).
matK: We used 20 µl reaction mix composed of 2.0 µl Buffer 10X, 0.8 µl MgCl2, 1 µl (~ 10 ng) primers, 4 µl Q-solution, 1.5 µl 1.25 mM l-1 dNTP, 0.2 µl (1 U) TAQ polymerase, 3 µl (~10 ng) DNA, then completed to volume (0.5 µl) with ultra-pure water. PCR reactions were conducted under the following protocol: 94 °C for 3 min + 30 cycles (94 °C for 1 min + 55 °C for 1 min + 72 °C for 2 min) + 72 °C for 7 min. Primers were Ac 12F and Ac 1290R (Miller and Bayer, 2001).
PCR products were sent for sequencing to Macrogen Korea. Assemblage and edition of the sequencing products were carried out in BioEdit v. 7.0.9 (Hall, 1999). The data were partitioned into three blocks according to the following gene regions: ETS, trnK, and matK. Each of the three partitions was aligned independently using MAFFT (Katoh et al., 2002; 2017) in the online server (https://mafft.cbrc.jp/alignment/server/). Alignments for each partition were generated using the default settings (gap opening penalty=1.53 and offset value=0.00). Finally, visual inspection and refinements were performed to optimize homology of the alignment.
Morphological characters
We assembled a morphological matrix with information compiled from the studies by Thompson (1980) and Gale and Pennington (2004) (Table 4). These characters and their states were assessed for structure, homology assumptions, and coding. To do so, we studied ~300 exsiccata studied at or else borrowed from the following herbaria: CICY, CIQRO, ENCB, FCME, GH, GUADA, MEXU, MO, UADY, UAMIZ, UCAM, UCOL, and US (acronyms as in Thiers, 2020, continuously updated). Thirty-five characters (both binary and multistate) were coded (Table 4). Of these, 26 were discrete, four were discrete numerical and, five were numerically continuous. In all cases, continuous characters which featured clear discontinuities in between were coded as distinct character states (for example character 9, leaflets size: microphyll (<15 mm long) or macrophyll (>20 mm long), character 15, corolla length: short (<5 mm long), or long (6.5-11 mm long), character 18, stamens length: short (<11 mm long), intermediate (15-20 mm long), and large (>25 mm long).
Table 4: Morphological characters and character states for the phylogenetic analysis of the genus Lysiloma Benth. based on Thompson (1980). Some characters are based on previous references, which are indicated in each case. The morphological matrix is included below the table.
| [01] Habit: 0 =tree, 1 =shrub, 2 =liana |
| [02] Stipule: 0 =foliaceous or subfoliaceous, 1 =espiniform |
| [03] Stipule, shape: 0 =ovate to widely ovate, 1 =linear (does not apply for the spiniform stipules) |
| [04] Stipules develop: 0 =absent, 1 =present |
| [05] Paraphyllidia: 0 =absent, 1 =present (Rico et al., 2008) |
| [06] Number of pinnae: 0 =1-2 pairs, 1 =3 or more pairs |
| [07] Number of leaflets per pinna: 0 =3-20, 1 =21 or more |
| [08] Mid vein: 0 =central, 1 =off center |
| [09] Size of leaflet: 0 =microphyll (<15 mm long), 1 =macrophyll (>20 mm long) |
| [10] Leaflet, shape: 0 =narrowly oblong, 1 =elliptic to widely elliptic |
| [11] Inflorescence, type: 0 =capitulum, 1 =raceme, 2 =fascicle |
| [12] Flower: 0 =sessile, 1 =pedicellate |
| [13] Number of flowers per head: 0 =few (<29), 1 =intermediate (30-35), 2 =many (>40) |
| [14] Bracteole, shape: 0 =spatulate, 1 =oblanceolate-linear, 2 =triangular-rhombic, 3 =cuneate |
| [15] Corolla, length: 0 =short (<5 mm long), 1 =long (>6.5-11 mm long) |
| [16] Stamens, connation: 0 =free, 1=joined into a short tube |
| [17] Stamens, number: 0 =few (<29) 1 =intermediate, (30-35), 2 =many (>40) |
| [18] Stamens, length (mm): 0 =short (<11 mm long), 1 =intermediate (15-20 mm long), 2 =large (>25 mm long) |
| [19] Fruit twisted in young state: 0 =absent, 1 =present |
| [20] Pod consistency: 0 =membranaceous, 1 =cartilaginous, 2 =chartaceous to crustaceous, 3 =coriaceous |
| [21] Pod, permanence: 0 =remain only one year, 1 =remains two or more year |
| [22] Pod, lateral sutures: 0 =always joined to the valves 1 =separating |
| [23] Craspedial pod dehiscence: 0 =tardily, 1 =early (does not apply to species without lateral sutures). |
| [24] Stipe of the pod, length: 0 =short (0.1- 1.5 cm), 1 =long (>2 cm). |
| [25] Pod, base: 0 =attenuate, 1 =obtuse 2 =truncate, |
| [26] Pod, shape of the apex: 0 =acute to narrowly acute 1 =obtuse o rounded-truncate to emarginated. |
| [27] Seeds leaving deep marks on the surface of the pod valves with ups and downs: 0 =absent, 1 =present |
| [28] Pod, exfoliation: 0 =absent, 1 =present |
| [29] Pod, venation: 0 =reticulate, 1 =parallel |
| [30] Funicule, shape: 0 =elongated and straight, 1 =sigmoid, 2 =almost absent, short |
| [31] Seed, shape: 0 =ovate to oblong, 1 =circular to quadrangular |
| [32] Seed, color: 0 =yellow to light brown, 1 =dark brown to black |
| [33] Pleurogram, relative size regarding the surface of the seed; 0 =less than 33, 1 =40-50%, 2 =more than 70% |
| [34] Seed areolate: 0 =absent, 1 =present (Rico et al., 2008) |
| [35] Cotyledon: 0 =not auriculate, 1 =auriculate |
| MATRIX (Polymorphism is indicated by *, and $, no apply is indicated by n). |
| Vachellia farnesiana 111n01*100000n0n0n10301nn0000120000 |
| Acaciella angustissima $011131*100012300200201n00000021110 |
| Acaciella villosa $011131*100012300200201201100021110 |
| Hesperalbizia occidentalis 00000110111000211220301n00010001111 |
| Mariosousa dolichostachya 111n011110011n1n0n10101n000001n0100 |
| Senegalia parviceps 211n001110000n0n0n10201n00000110010 |
| Lysiloma acapulcense $000001110011110101121010*011001110 |
| Lysiloma auritum 00000111100111101111210100011001110 |
| Lysiloma candidum 00000n00001000001111210100011000110 |
| Lysiloma divaricatum 00000011000000001011210100011000110 |
| Lysiloma latisiliquum 000000*10000001010$1210010010000110 |
| Lysiloma microphyllum 00000011000000001011210100011000110 |
| Lysiloma sabicu 000001*0011010011001010011111000110 |
| Lysiloma tergeminum 0000010011200000100121010111100*110 |
| Lysiloma watsonii 10000111100110101111210100011001110 |
Phylogenetic analyses
Morphological analysis
We prepared a matrix composed by morphological data only (Table 4) that was analyzed with a Maximum Parsimony Analysis done with NONA v. 2.0 (Goloboff, 1993) through the Winclada v. 1.00.08 (Nixon, 2002) shell. Non-parsimony informative coding characters were deactivated. Informative characters were considered unordered and given the same weight (Fitch Parsimony). To identify maximally parsimonious topologies, we performed a ratchet algorithm analysis with 5000 iterations, 10 trees held at each iteration, whereas 10% of the characters were sampled in each iteration (mult* 10000, ho/10; max*). Clade support was assessed with 1000 iterations of bootstrap. The topologies retrieved are shown here only for morphological characters, but our complete set of results is available from the corresponding author upon request.
Phylogenetic analyses
Morphological analysis
There has been much debate over the merits of different algorithms for phylogenetic inference (Rindal and Brower, 2010). However, parametric Bayesian methods have become very popular in molecular phylogenetics due to the availability of user-friendly software implementing sophisticated models of evolution (Nascimento et al., 2017). These methods have the advantage of including nucleotide evolutionary models and a solid statistical framework. In the present study, we use maximum parsimony as an exploratory analysis, obtaining topologies (not shown) that are highly congruent with those resulting from Bayesian analyses.
We assembled several DNA matrices, one composed only of rDNA-ETS sequences that included 112 taxa and account for most of the major clades in tribes Ingeae and Acacieae that have been identified in recent analyses (e.g. Brown et al., 2008, 2011, Iganci et al., 2015, Ferm, 2018). The analysis was carried out with MrBayes version v. 3.2.7 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and Tracer v. 1.6 (Rambaut et al., 2014).
This analysis was designed to test the monophyly of Lysiloma as well as the position of the genus within Ingeae + Acacieae. The matrix is available from the corresponding author upon request. Except for the sequences of Lysiloma, Zapoteca H.M. Hern., and some members of the Pithecellobium alliance, most of the sequences used were retrieved from GenBank (GenBank, 2020; Table 2). The individual matrices were integrated with our data and completed with GenBank sequences to match the composition of the morphological matrix.
Molecular analyses
We concatenated the three molecular alignments and morphological data in a matrix and analyzed it under the Bayesian Inference paradigm using MrBayes v. 3.2.7 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and Tracer v. 1.7.1 (Rambaut et al., 2014).
Each partition was treated as independent and associated with its own model. Our analyses were performed with the default parameters of the software, except for the number of generations, which were five million. Two independent threads were run. Convergence was assessed with both MrBayes v. 3.2.7 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and Tracer v. 1.7.1 (Rambaut et al., 2014). Posterior Probabilities (PP) of <0.95 were considered weakly supported, whereas PP of 0.95-1.0 were deemed as strongly supported. Missing data were coded as “?” in the concatenated matrix. To assess the best evolutionary model for all the molecular matrices, we used jModelTest v. 2.1.7 (Guindon and Gascuel, 2003; Darriba et al., 2012). In each case, three substitution schemes were used; the search criterion was a maximum likelihood tree estimated with the “best” option of the software. The selected model was suggested by the Akaike Information Criterion (AIC). For the larger ETS matrix, the best model was GTR + G, as well as for the smaller matrices (matK, trnK and ETS).
Combined analyses
We concatenated the three molecular alignments and morphological data in a matrix and analyzed it under the Bayesian Inference paradigm using MrBayes v. 3.2.7 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and Tracer v. 1.7.1 (Rambaut et al., 2014).
Each partition was treated as independent and associated with its own model. Our analyses were performed with the default parameters of the software, except for the number of generations, which were five million. Two independent threads were run. Convergence was assessed with both MrBayes v. 3.2.7 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) and Tracer v. 1.7.1 (Rambaut et al., 2014). Posterior Probabilities (PP) of <0.95 were considered weakly supported, whereas PP of 0.95-1.0 were deemed as strongly supported. Missing data were coded as “?” in the concatenated matrix. To assess the best evolutionary model for all the molecular matrices, we used jModelTest v. 2.1.7 (Guindon and Gascuel, 2003; Darriba et al., 2012). In each case, three substitution schemes were used; the search criterion was a maximum likelihood tree estimated with the “best” option of the software. The selected model was suggested by the Akaike Information Criterion (AIC). For the larger ETS matrix, the best model was GTR + G, as well as for the smaller matrices (matK, trnK and ETS).
Fossil calibration and diversification times
A Bayesian analysis to estimate divergence times was conducted using the ETS matrix under the uncorrelated lognormal relaxed molecular clock approach implemented in the program BEAST v. 1.10 (Suchard et al., 2018). The main reason to use this marker is the high number of accessions available in GenBank (Brown et al., 2008; Murphy et al., 2010; Ferm. 2018).
In each BEAST 1-10 run, we used pure-birth (Yule) tree prior, and a Monte Carlo Markov chain (MCMC) of 25,000,000 generations, sampling every 1000 generations, with parameters sampled every 1000 generations. For this matrix, the nucleotide substitution model was GTR + I + G using the AIC criterion with jModelTest v. 2.1.6 (Posada, 2008).
Tracer v. 1.7.1 (Rambaut et al., 2014) was used to assess effective sample sizes (ESS >200) for all estimated parameters. We used TreeAnnotator v. 1.8.2 (part of the BEAST package) to discard 10% of the saved trees as burn-in and to combine trees. Maximum clade credibility trees with mean node heights were visualized using FigTree v. 1.4.2 (Rambaud, 2014). We report highest posterior densities intervals, the interval containing 95% of the sampled values.
Three fossils were used to calibrate the divergence dating analysis. The first one was Lysiloma mixtecana (Magallón-Puebla and Cevallos-Ferriz, 1993) assigned to the crown group of Lysiloma at 28.4 million years (myr). The second point was based on pollen assigned to Calliandra Benth. (Caccavari and Barreda, 2000), with 16 myr and referred to the core group of Calliandra, whereas the third one was the tree root with 45 Ma, a date taken from the fossil pollen of the most recent common ancestor assigned to Ingeae and Acacieae (Simon et al., 2009). All fossils were defined as minimum age constraints and implemented in the dating analysis as a lognormal statistical distribution. We choose lognormal distribution because it is appropriate for calibrations derived from fossils. A log normal density distribution to calibration points allows for uncertainty associated with a fossil representing a minimum age where the clade in question could have evolved earlier but not later than the age of the fossil. The maximum clade credibility tree (MCC) was visualized using FigTree v.1.4.2 (Rambaud, 2014) and the means and 95% higher posterior densities (HPD) were obtained.
We carried out a second estimate of divergence times analysis intended to evaluate the origin of the genus without any temporal constraint. For this reason, we did not include the fossil of Lysiloma mixtecana, but we used the other two fossils plus Acaciapollenites myriosporites (Cookson) Mildenhall (23 Ma) (Macphail and Hill, 2001), which was assigned to the Acacia s.s. clade.
For this analysis we had to implement a normal prior with a stdev=2 for the older date (Ingeae and Acacieae 45 Ma) and the other fossils were assigned with a gamma prior and stdev=2 distribution, because these distributions allowed us to obtain the best results for the effective sample size (ESS). These parameters have also been implemented in other studies (Gustafsson et al., 2010; Chomicki et al., 2015; Pérez-Escobar et al., 2017).
Results
Morphology
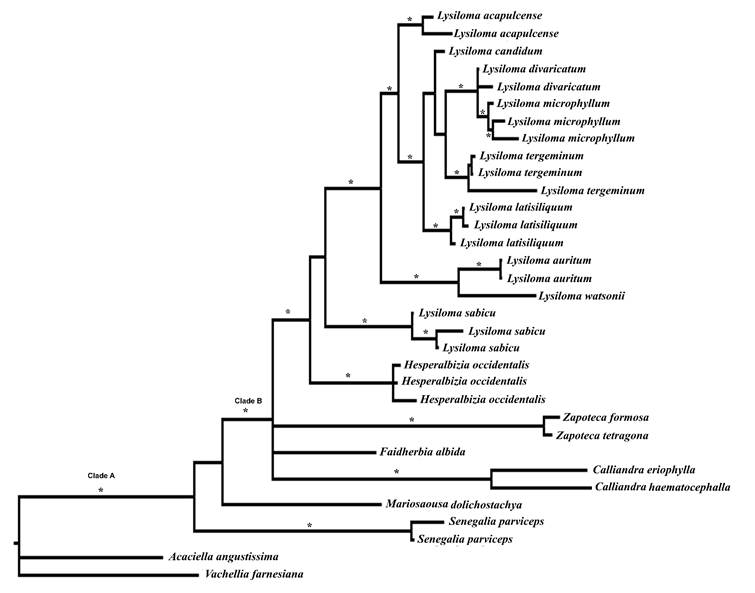
The parsimony analysis based on morphological characters yielded eight most parsimonious trees (L=63, CI=69, RI=80). Figure 2 shows the strict consensus tree, where three nodes have no support and collapse. Vachellia farnesiana, upon which the cladograms are rooted, is followed by a poorly supported grade that includes Senegalia parviceps, Mariosousa dolichostachya, Acaciella spp. plus a clade containing Lysiloma and Hesperalbizia. The clade including Acaciella Britton & Rose as a sister group of Lysiloma and Hesperalbizia has moderate bootstrap support (77%) and is held together by four synapomorphies. Acaciella has a strong bootstrap support (97%) and is supported by two synapomorphies. The clade Hesperalbizia and Lysiloma (clade A) also features a high bootstrap support (91%) and is supported by four synapomorphies. Hesperalbizia is supported by one synapomorphy. On the other hand, Lysiloma (clade B) is monophyletic and has a low bootstrap support (68%); this clade features five morphological synapomorphies. Internally, Lysiloma is poorly resolved; there are three terminal taxa; L. auritum (Schltdl.) Benth., L. acapulcense and L. watsonii Rose and clade C. The last clade lacks bootstrap support, but it is held together by four reversions. It is composed of L. latisiliquum and two clades. The first, including L. divaricatum + L. microphyllum Benth. with a low bootstrap support (53%), and the second composed of three species with moderate bootstrap supported (77%): L. candidum Brandegee as a sister group of L. tergeminum Benth., and L. sabicu.
Clade C, which is composed by L. watsonii and the clade L. acapulcense and L. auritum is strongly supported by the bootstrap (94%). Clades D (L. candidum plus the clade L. sabicu + L. tergeminum), E (L. sabicu + L. tergeminum), and F (L. divaricatum + L. microphyllum) lacked significant bootstrap support (<50%).
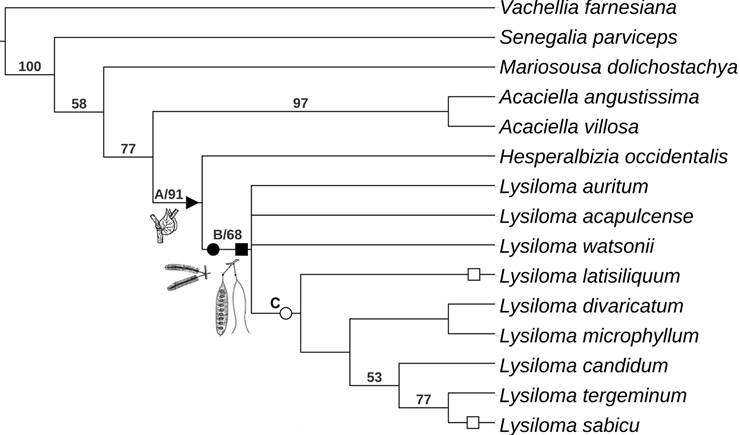
Figure 2: Results of parsimony analysis of the genus Lysiloma Benth. based on morphology (value below the node, bootstrap support, simbols: A, clade Hesperalbizia and Lysiloma; B, clade Lysiloma; C, species with inflorescence racemose; black triangle (u), synapomorphy: stipule developed, black circle (˜) and black square (¢): synapomorphies for inflorescence racemose and pod craspedial; white circle (™): inflorescence capitate (reversion), and white square (£): pod indehiscent (reversion).
Molecular data
Figure 3 shows the result of the Bayesian analysis (rDNA-ETS). In this analysis, some of the genera segregated from Acacia Mill. such as Acaciella, Mariosousa Seigler & Ebinger, Senegalia Raf., and Vachellia Wight & Arn. are poorly represented. However, this large analysis evaluates the relationship of the genus Lysiloma in a more general context (see Brown et al., 2008, for a complete list of the genera of the Acacieae+Ingeae tribes). Acaciella, Senegalia, Mariosousa comprise a basal grade. The clade A (PP=1.0) is formed for two clade: Calliandra is the sister group Zapoteca and both are the sister group of Ingeae, including Acacia (B Clade, PP=1.0). This clade has two subclades: the clade Viguieranthus sensu Rodriguez de Souza et al. (2016) including Faidherbia A. Chev., Sanjappa É.R. Souza & M.V. Krishnaraj, Thailentadopsis Kosterm., and Viguieranthus Villiers (PP=0.95) and clade C. This clade included Cojoba Britton & Rose as sister group of Lysiloma and Hesperalbizia, and clade D (PP=0.99). This last clade includes four subclades whose relationships are unresolved, namely the New World Ingeae p.p. clade, which is strongly supported (PP=1.0), the poorly supported (PP=0.76) Old World Ingeae clade, including Acacia, and lastly, the Pithecellobium clade (part of the New World Ingeae), which is strongly supported (PP=1.0).
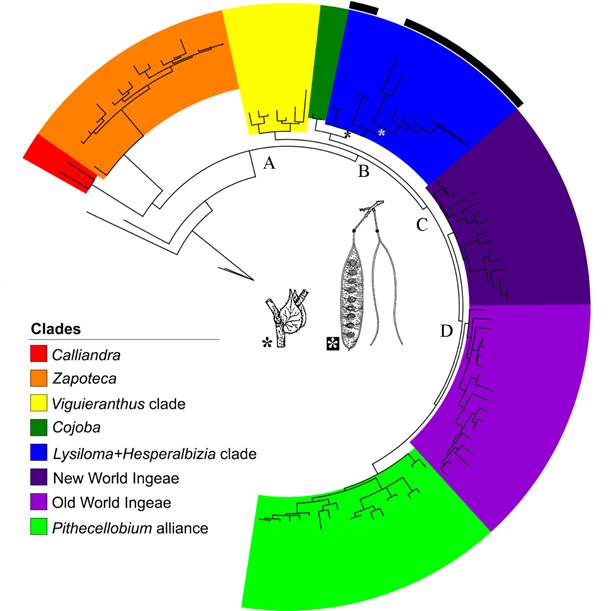
Figure 3: Results of Bayesian analysis of the genus Lysiloma Benth. based on a large molecular matrix of DNA-ETS (Clade A: core Ingeae, including Acacia Mill. clade B: clade A- clade Calliandra, and Zapoteca; clade C: Cojoba, sister group of Lysiloma and Hesperalbizia, and Ingeae p.p. including Acacia, Clade D: New World Ingeae p.p., Old World Ingeae and Acacia, and Pithecellobium alliance. Important synapomorphies: stipule developed for Lysiloma + Hesperalbizia, and pod craspedial for Lysiloma.
Combined molecular and total evidence analyses
From all regions analyzed, we found ETS to be the most informative: 26 taxa, 482 characters, from which 44.73% were informative, followed by trnK, 21 taxa, 999 characters, 5.4% informative, and by matK, 18 taxa, 767 characters, 3.65% informative. The results of the combined molecular analysis and those of the total evidence analysis are very similar; thus, we only show the second in Figure 4. At the base of the tree is a polytomy conformed by Vachellia farnesiana (root), Acaciella, and then it follows a clade with all the other taxa (Clade A). This polytomy is the sister group of a grade with Senegalia parviceps, Mariosousa dolischotachya, and clade B. This last clade included four subclades without resolution among them, namely Calliandra spp., Faidherbia, Zapoteca spp., and finally a clade with Lysiloma and Hesperalbizia (Clade C). In the combined molecular analysis as well as each individual analysis (ETS, matK, trnK), there is no resolution for Hesperalbizia occidentalis, Lysiloma sabicu and the core of Lysiloma. However, in the total evidence analysis, Hesperalbizia was recovered as sister group of Lysiloma with low support.
Diversification of the genus Lysiloma
The exclusion of the Lysiloma fossil from the molecular clock analyses does not result in relevant differences in chronology. Hence, the results are not contingent upon the restriction imposed by the inclusion of this fossil and associated date.
Figure 5 shows the result molecular clock analyses. The divergence of Lysiloma and Hesperalbizia (L1) was estimated at about 28.22 myr (95% HPD: 19.5-36.25) in the beginning of the Oligocene. The second node (L2) corresponds to the divergence of Lysiloma and was estimated at about 29.86 myr (95% HPD: 28.64-39.13). Node L3, the divergence of the Lysiloma core, was estimated at about 22.65 myr (95% HPD: 15.78-28.33) in the beginning of the Miocene.
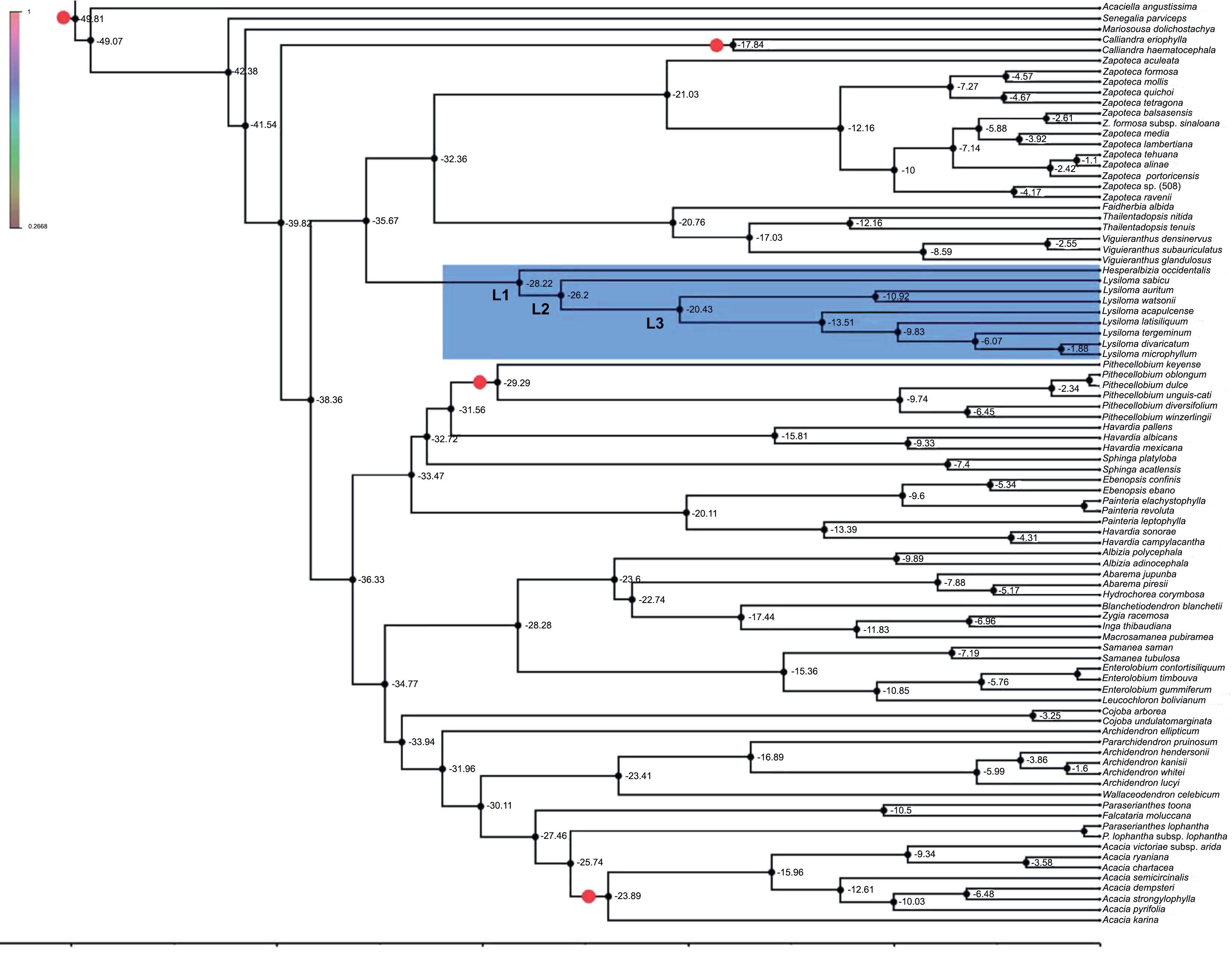
Figure 5: Chronogram of Lysiloma, and Hesperalbizia and other related taxa from the tribe Acacieae and Ingeae (Caesalpinioideae) based on ETS data. Divergence times are shown using the computer program BEAST v. 1.10. The calibration nodes 1 (28.4 myr), and 2 (16 myr), are marked by dots based on the fossil records. The root of the tree was set to no more than 45 myr.
Discussion
Phylogeny reconstruction is a crucial aim of contemporary systematics. The success of phylogenetic inference can be measured in terms of resolution, support, and accuracy (Wortley et al., 2005). Many studies suggest that increasing sequence data is a better way to improve support, resolution, and accuracy of the phylogenetic trees (Rokas and Carroll, 2005) at any level; the ordinal level (Li et al., 2019), family level (Stull et al., 2015), generic level (Cardoso et al., 2015; Stull et al., 2015), and at the species level (Nicholls et al., 2015). We consider that additional DNA regions must be explored to test and improve the resolution of topology, particularly regarding Lysiloma and Hesperalbizia. The morphological analysis (maximum parsimony) confirms the monophyly of Lysiloma and its close relationship with Hesperalbizia, in concordance with two previous molecular analyses, which include at least one accession of each genus (Brown et al., 2008; Iganci et al., 2015). However, the current morphological analysis does not allow to recover a complete picture of the relationships of Lysiloma and Hesperalbizia with other members of tribes Acacieae and Ingeae, because only a few taxa were included.
Morphological analysis
In a previous morphological analysis that included many members of the tribe and in which 75 characters were used (Grimes, 1995), a different picture of the relationships of Lysiloma and Hesperalbizia was retrieved. Grimes (1995) found that Lysiloma is most closely related to the Pithecellobium alliance, along with Faidherbia, Senegalia, and Vachellia. However, Hesperalbizia is more related to Samanea (Benth.) Merr., and Pseudosamanea Harms. The main explanation for this result is the unorthodox way of coding some characters by the abovementioned author (e.g. characters 51 to 58 are derived from the fruit and represent one or two, probably strongly correlated characters).
According to our morphological analysis, Lysiloma has five synapomorphies (Fig. 2, characters 16, 18, 20, 21, and 29). The fact that some of these characters states are synapomorphies may be related to the sampling strategy. Hence, if more taxa of the tribe were included, some would most likely become homoplasic. For example, the character “fruits remaining united to the mother plant for long time” is found in Albizia Durazz. (not included in the current analysis). In addition, the character “prominent foliaceous stipules” a synapomorphy for Hesperalbizia+Lysiloma is present in Albizia chinensis (Osbeck) Merr.
Molecular and total evidence analyses
In a more general context, three previous phylogenetic analyses have been published with abundant accessions of ITS, and/or ETS (Brown et al., 2008; Rodriguez de Souza et al., 2013; Iganci et al., 2015). In concordance with those, our large (rDNA-ETS, Fig. 2) analysis supports a basal position for the segregates of Acacia, namely Acaciella, Mariosousa, Senegalia, and Vachellia as well as for the Lysiloma+Hesperalbizia clade. It also supports the monophyly of the Pithecellobium alliance and the clade Viguieranthus as a sister group of Zapoteca (Rodriguez de Souza et al., 2016). None of these earlier phylogenetic analyses support the relationship of Lysiloma and Hesperalbizia found by Grimes (1995), nor support the relationship of Hesperalbizia with Albizia (Rico Arce et al., 2008). In the current analysis some clades are strongly supported (e.g. Zapoteca, Cojoba, and the Pithecellobium alliance (PP >98%)), but not the Lysiloma+Hesperalbizia clade. The basal clade of the current tree (Fig. 2) is a mixture of elements restricted to the New World (Acaciella, Mariosousa, and Zapoteca), others from the Old Word (Faidherbia), as well as elements from both regions (Calliandra+Afrocalliandra É.R. Souza & L.P. Queiroz, and Senegalia). There are three clades with no resolution which reveal geographic coherence: The New World Ingeae (e.g. Abarema, Inga Mill., Samanea, Enterolobium Mart., etc.), the Old World Ingeae (e.g. Archidendron F. Muell., Paraserianthes I.C. Nielsen, Wallaceodendron Koord., etc.), plus Acacia s.s., and the Pithecellobium alliance from the New World.
Regarding the internal relationship of the genus, Lysiloma latisiliquum and L. sabicu, the two taxa bearing fruits with persistent lateral sutures are not retrieved as a clade; instead, the two species are nested in widely diverging clades, pointing to an independent evolution of this fruit type. Our results suggest that the distribution of L. sabicu in the Antilles can be explained by an old vicariance process but in the case of L. latisiliquum, it requires long-distance dispersal (from mainland Mexico to south Florida, Bahamas, Turks and Caicos Islands, Cuba and Haiti). A fact that still has no explanation is why L. sabicu has been less successful in its expansion in the Antilles compared to L. latisiliquum, considering the long occupation time of the lineage in the area.
Lysiloma taxa with capitate inflorescences and those with racemose inflorescences constitute polyphyletic assemblages. In summary, we found no support for Thompson’s infrageneric informal classification (1980). This was already proposed by Barneby and Grimes (1996), who did not find any reason to maintain such a classification and proposed that character states observed in L. latisiliquum and L. sabicu probably evolved independently. The evolution of fruit dehiscence in Lysiloma is of ecological interest, because indehiscent fruits, which are buoyant and probably an apomorphic condition, have most likely allowed the genus to invade the West Indies.
In our analysis, we did not find a clear biogeographical pattern with the three species distributed in the Nearctic region (the southwestern coast of USA, and Mexico): L. candidum Brandegee, L. microphyllum (partially) and L. watsonii, because they are not closely related, suggesting the genus invaded the Nearctic region thrice. However, a structurally and geographically coherent clade recovered in both the morphological and total evidence analyses was found to occur along the Pacific coast of Mexico: L. candidum, L. tergeminum, and L. divaricatum. One of the morphological features of this clade are the leaves with few pinnae. Therefore, diversification in Lysiloma seems to have followed an isolation by allopatry pattern, whereby sister taxa are currently allopatric and sympatric taxa are not closely related. Thus, Lysiloma latisiliquum and L. sabicu, which are sympatric in the West Indies, belong to different clades in the genus, whereas the sister pair of L. watsonii and L. auritum occur one each on one of the opposing drainages of Mexico, the former on the Pacific slope, the latter on the Gulf slope.
Lysiloma sabicu is the most distinct taxon within the genus. This taxon shares with Hesperalbizia leaves with few pinnae with a few large leaflets, which can imply a morphological transition. It is possible that additional evidence will eventually increase the support for a sister group relationship of Hesperalbizia and Lysiloma. In the total evidence analysis (Bayesian), Hesperalbizia is the sister group of Lysiloma, but the relationship is poorly supported (PP <0.95%).
The advantages of including Hesperalbizia in the genus Lysiloma are similar to those of creating a monotypic genus for Lysiloma sabicu. Any decision here, regarding the generic boundaries of Lysiloma and Hesperalbizia, should be well thought-out and meet the mandatory rule of monophyly, as well as the secondary criteria proposed by Backlund and Bremer (1998): support, diagnosability, maximum informativity, and stability. There are morphological differences in the most distinctive character of Lysiloma as compared to Hesperalbizia; for example, the fruit in Lysiloma exfoliates, eventually revealing the pale brown endocarp (not in Hesperalbizia). Another difference is related to flower number: fewer with longer petals in Hesperalbizia. Seeds are also different (lens-like, pale brown, and areolate in Hesperalbizia), as opposed to ovate, oblong to elliptic and dark brown to black in Lysiloma. Thus, here we argue that until more data become available, it is best to retain Lysiloma in its present circumscription.
Despite Lysiloma being a small genus, it shows a complex evolutionary structure. The phylogenetic molecular analysis points at the homoplasic evolution of many morphological characters, which strongly suggest that different lineages were modeled by similar ecological pressures. Other examples of a similar pattern are the lizards of the genus Anolis Daudin, where in the case of Caribbean species, the evidence strongly supports the hypothesis of repeated and independent development of similarly shaped body on each island (Losos, 2001). In the case of flowering plants, the genus Manihot Mill. is another good example of convergent evolution (in this case, growth forms) (Cervantes-Alcayde et al., 2015). Both examples are of speciose lineages with 400 and 100 species, respectively. Because of this phenomenon (repeated evolutionary convergence), the classifications based only on morphology may be misleading, being not natural (monophyletic), but polyphyletic or paraphyletic. Moreover, although these two examples may suggest that repeated evolutionary convergence is characteristic of diverse lineages, our data of Lysiloma show that even small genera with more restricted distributions can also show this pattern. A similar pattern of evolution could be found in other members of the tribe Ingeae, involving some Old World Albizia species and New World Albizia species associated with flooded forests, all the species of the genus Hydrochorea Barneby & J.W. Grimes, and the two species of Balizia Barneby & J.W. Grimes section Balizia. All these lineages are subject to environmental pressures (seasonally flooded environments) and present indehiscent, lomentiform fruit.
In addition, there is a long-standing argument on the position of Lysiloma microphyllum relative to L. divaricatum, to whose synonymy it has been relegated. Thompson (1980) recognized both as distinct, albeit morphologically very similar. However, Barneby and Grimes (1996) and Gale and Pennington (2004) were unable to identify consistent characters to separate them. Upon evaluating morphological features in herbarium specimens, we did not find differences in character states between both taxonomic concepts. Furthermore, molecular data do not support both taxa as separate entities, as their ETS, matK, and trnK sequences are almost identical. Neither is there a geographical discontinuity in their distributional ranges (Thompson, 1980). Thus, here we conclude that L. divaricatum and L. microphyllum should be treated as the same species (see Gale and Pennington, 2004; Andrade and Sousa, 2012).
Lysiloma acapulcense requires special consideration. As presently circumscribed, this taxon includes a long list of synonyms, reflecting the fact that it is an extremely polymorphic and widely distributed species. A review of the fruit of the types allows us to recognize at least two additional morphotypes: one characterized by very long and thin fruits (Lysiloma jorullense Britton & Rose), and a second morph bearing fruits with a broad base (Lysiloma platycarpa Britton & Rose). Thus, this species requires a taxonomic re-evaluation.
Diversification of the genus Lysiloma
The divergence between Lysiloma and Hesperalbizia (Fig. 5, L1) was estimated at 28.2 myr at the beginning of the Oligocene (Fig. 5, Table 4). This date coincides with a drastic and abrupt decrease in temperature and humidity at the beginning of the Oligocene (Galeotti et al., 2016). The second node (L2) corresponds to the divergence of the Lysiloma estimated at 26 myr, coinciding with two important facts, a low global temperature and humidity as well as a proximity between the Caribbean Arc and the American continent through the Yucatan peninsula during the Oligocene (Pindel and Kennan, 2009). An important lineage divergence occurred in the upper Miocene, whereas additional minor ones happened below this node (N3) all along the Miocene, which agree with Becerra’s (2005) findings regarding the evolution and divergence of Bursera (Burseraceae) in the dry forests.
We can assume that the history of Lysiloma seems to be tightly related to the last 30 to 5 myr corresponding with the formation of the main orographic systems in Mexico, such as the Sierra Madre Occidental and later the Neovolcanic belt and the dry forest (Gómez-Tuena et al., 2007). The last uplift of the Sierra Madre Occidental occurred between 34 and 15 myr, whereas the Neovolcanic axis was formed in several stages along a west-east progression that started in the west (Sosa et al., 2018). The uplift of these two orographic systems was presumably responsible for the climatic conditions necessary (high temperature and low humidity) for the development and maintenance of the dry forests (Sosa et al., 2018).
Conclusion
Lysiloma and Hesperalbizia are sister genera, albeit with low clade support, with Lysiloma often resolved as paraphyletic with respect to Hesperalbizia. More data are necessary to confirm the generic status of Lysiloma. This sister clade is early branching within the clade comprising the rest of the tribe Ingeae. Indeed, we estimate the stem age of this sister clade to average about 32 Ma. In addition, our analysis shows that, although Lysiloma is a small genus, it shows a complex evolutionary structure that may be modeled in different lineages of the genus by the same ecological pressures, suggesting that the phenomena of convergence and/or parallel evolution occur regardless of species richness. One consequence of this phenomenon (repeated evolutionary convergence) is that classifications based on morphology are not necessarily natural (monophyletic), but instead, a conglomerate of polyphyletic or paraphyletic taxa. This applies not only to morphology, but also to patterns of geographic distribution: unrelated species can occupy the same distribution.
Regarding the divergence time, the history of the genus Lysiloma and Hesperalbizia begins in the Oligocene and diversifies into the beginning of the Miocene. This time frame coincides with low global temperatures, a proximity between the Caribbean Arc and the American continent through the Yucatan peninsula, and the uplift of the Sierra Madre Occidental and later of the Neovolcanic axis. Presumably, these conditions were responsible for the climatic conditions associated with the development and maintenance of the dry forest in Mexico (Sosa et al., 2018), the habitat in which Lysiloma diversified.











 nueva página del texto (beta)
nueva página del texto (beta)