Introduction
The direct extraction of microorganism’s nucleic acids material from the soil or any other environmental sample has become a useful tool to identify microorganisms that cannot be cultivated in laboratory conditions. The direct extraction of DNA reveals genotype diversity in different microbial ecosystems (Sagova-Mareckova et al., 2008; Islam, Sultana, Melvin-Joe, Cho, and Sa, 2012). Increased yields of DNA from soils have been obtained employing different physical-mechanical methods, e.g., mechanical bead beating (Cullen and Hirsch, 1998), sonication to lyse microbial cells (Krsek and Wellington, 1999), manual fragmentation using a mortar and pestle (Devi et al., 2015). However, those treatments can reduce DNA to fragments <10 kb (Liesack and Stackebrandt, 1992; Zhou, Bruns, and Tiedje, 1996). These small DNA fragments presents some drawbacks for the analysis of microbial communities, because of chimeric products formation with small template DNA in the PCR amplification process (Hugenholtz and Huber, 2003).
Different methods for direct DNA extraction have been proposed for environmental samples, e.g. mannitol-based methods, polyethylene glycol (PEG/NaCl) method (Fatima, Pathak, and Rastogi, 2014), guanidinium isothiocyanate-based lysis (Hill et al., 2015), and bead beating-phosphate lysis (Guerra, Beule, Lehtsaar, Liao, and Karlovsky, 2020). However, some microbial cells may remain strongly bonded to soil particles, this process difficult the DNA extraction with high yields, mainly in soil with high content of clay or organic matter (Zhou et al., 1996). Additionally, the DNA extracted from the soil regularly co-extracts organic substances, e.g., humic or fulvic acid, which hinders DNA detection and quantification (Zipper et al., 2003). This organic substance can inhibit DNA polymerase in the PCR amplification (Ogram, 2000), and interferes in specific process of PCR like restriction enzyme digestion, transformation efficiency, and DNA hybridization (Tsai and Olson, 1992). So, the removal of the humic and fulvic acids co-extracted in the direct extraction of DNA from the soil is an important step before the amplification of DNA by PCR (Steffan, Goksøyr, Bej, and Atlas, 1988).
Extraction of cells from the soil is another way of studying microorganisms in their natural environments (Cho, Lee, Cheol, Cho, and Kim, 1996) producing high purity of the DNA extract, but a large amount of soil is needed, the technique is very complex and time consuming and some bacteria cannot be extracted (Courtois et al., 2001). The direct extraction method (SDS-based), however, results in a 92-99% recovery of DNA from soil bacteria and crude DNA yield, agreeing reasonably well with a yield based on microbial counts (Zhou et al., 1996). Most proposed extraction methods were developed for a limited number of soil types and their efficiency in other environments is unknown (Alm and Stahl, 2000; Hurt et al., 2001). Some extraction method has been developed for alkaline soils (Sorokin et al., 2001; Narayan, Jain, Shah, and Madamwar, 2016; Gupta, Manjula, Rajendhran, Gunasekaran, and Vakhlu, 2017), but we are not aware of any studies done with soils with high pH, and large electrolytic conductivity (EC) and organic matter content.
The former lake Texcoco has unique conditions of salinity and alkalinity. It is located in the valley of Mexico City at an altitude of 2240 m above the average sea level with a mean annual temperature of 16 °C and precipitation 705 mm (Dendooven et al., 2010). The soil of the former lake has partly been drained to reduce the excess of salt and irrigated with sewage effluents, which in turn affects soil characteristics (Luna-Guido et al., 2000). Natural spatial variation, drainage with effluents for different lengths of time, and introduction of vegetation has created soils in the former lake Texcoco with different pH and salinity (Luna-Guido et al., 2000; Gutiérrez-Castorena, Stoops, Ortiz, and López, 2005). The undrained soil present variable pH ranging from 9.8 to 11.7, electrolytic conductivity from 22 to 150 dS m-1 and contains a high concentration of sodium (exchangeable sodium percentages >76% and sodium adsorption ratio >103 Mm) (Beltrán-Hernández et al., 1999).
The aim of this study was to evaluate the extraction of DNA from saline-alkaline allophanic soils with large organic matter content and develop a simple and improved method giving large DNA yields with sufficient large fragments so that the microbial community in this specific environment could be studied more in detail. The commercial extraction kit Power Soil DNA (Mo Bio™ Laboratories, Inc.) was used as a reference technique. The method reported here was tested on four soils with EC between 39.9 dS m-1 and 0.8 dS m-1, pH between 10.9 and 6.3, and organic matter content between 30 and 12.7 g C kg-1 soil. The quality of the extracted DNA was tested by PCR amplification of the bacterial 16S rDNA marker gene, as polymerase is sensitive to contamination with humic acids.
Materials and Methods
Soil sampling
Three different locations with different EC and pH in the former Lake Texcoco (state of Mexico, Mexico) were sampled. The first site had a pH of 10.9 and EC of 39.9 dS m-1 (considered Texcoco-1), the second site had a pH of 9.4 and EC of 9.8 dS m-1 (considered Texcoco-2), and the third location had a pH of 8.8 and EC of 2.6 dS m-1 (considered Texcoco-3). More details of the characteristics of the Texcoco soil can be found in Dendooven et al. (2010). The Acolman soil, which served as a control, was sampled near the ex-convent of Acolman in the State of Mexico, Mexico (Table 1). More details of the characteristics of the Acolman soil can be found in Betancur-Galvis, Alvarez, Ramos, and Dendooven (2006).
At each sampling site (n = 4), the 0-10 cm layer of three 400 m2 areas were collected 20 times each. The soil collected from each area was pooled separately so that 12 soil samples were obtained. A 100 g sub-sample was taken from each soil sample and stored at -80 °C until extracted for DNA.
Table 1: Characteristics of soil samples used in this study.
Soil |
Location |
pH |
Electrolytic conductivity |
Carbon |
Particle size distribution |
USDA textural classification |
|||
Organic |
Inorganic |
Clay |
Silt |
Sand |
|||||
dS m-1 |
- - - - - - - - g kg-1 soil - - - - - - - - |
||||||||
Texcoco 1 |
N 19°30'05'' W 98°59'15'' |
10.9 |
39.9 |
30.3 |
7.5 |
444 |
167 |
399 |
Clay |
Texcoco 2 |
N 19°29'80'' W 98°58'01'' |
9.4 |
9.8 |
26.8 |
8.4 |
312 |
55 |
634 |
Sandy clay loam |
Texcoco 3 |
N 19°29'46'' W 98°58'05'' |
8.8 |
2.6 |
12.7 |
12.2 |
239 |
40 |
722 |
Sandy clay loam |
Acolman |
N 19°38 W 98°55 |
6.3 |
0.8 |
19.0 |
0.6 |
440 |
300 |
260 |
Clay |
Soil characterization
The collected soil samples were air-dried in the shadow for 7 days, grinded afterward and sieved at 5 mm-mesh. Subsequently, the soil was characterized (Table 1). The EC was measured in a saturated solution extract (Conde et al., 2005). Soil pH was measured in 1:2.5 soil-H2O suspension using a glass electrode (Thomas, 1996). The total C in soil was determined by oxidation with potassium dichromate and trapping the evolved CO2 in NaOH, followed by titration with 0.1 M HCl (Amato, 1983). Inorganic C in soil was determined by adding 20 mL 1 M HCl solution to 1 g air-dried soil and trap CO2 evolved in 20 mL 1 M NaOH and then titrate. The organic C was defined as the difference between total and inorganic C. Total N was measured by the Kjeldahl method (Bremner, 1996) and soil particle size distribution by the hydrometer method (Gee and Bauder, 1986).
Extraction of total soil DNA
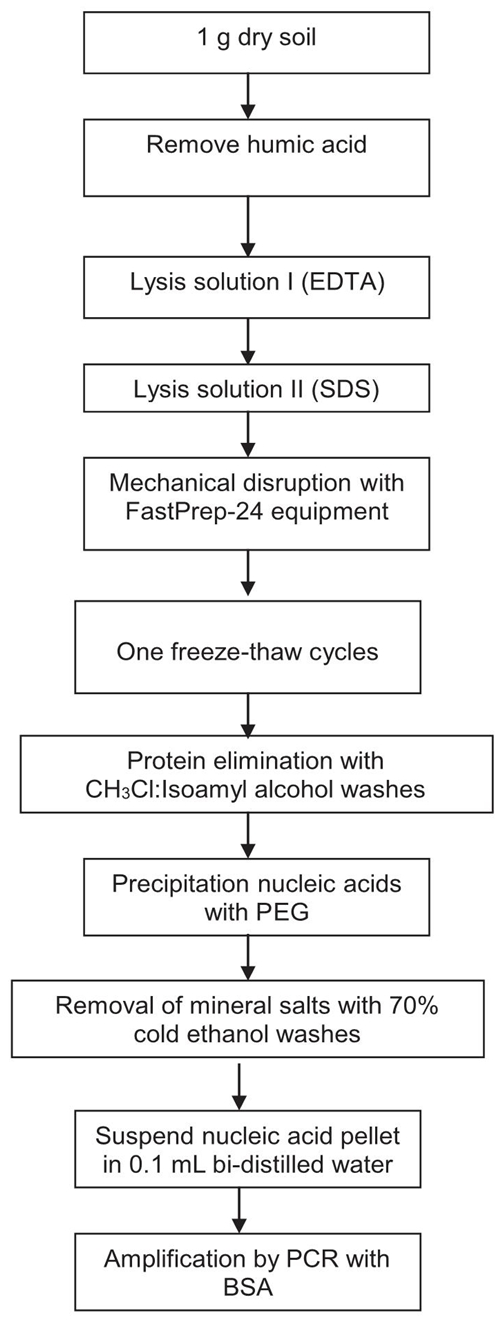
The total DNA from soil was extracted using a modified method (Tsai and Olson, 1991; Erb and Wagner-Döbler, 1993; Guo, Sun, Harsh, and Ogram, 1997; Valenzuela-Encinas et al., 2008) (Figure 1). The composition and concentration of the solutions used for DNA extraction are shown in Table 2. A sample of 1 g soil was added to 15 mL conical centrifuge tubes containing 10 mL of sodium pyrophosphate solution for humic acid removal (Ceja-Navarro et al., 2010), vortexed for 1 min, incubated for 10 min at room temperature, and centrifuged for 10 min at 3000 × g. The supernatant was decanted, and the step with the sodium pyrophosphate solution was repeated four times. Soil pellet was re-suspended in 10 mL of sodium phosphate buffer solution (PB buffer), vortexed for 1 min, incubated for 10 min at room temperature, and centrifuged for 10 min at 3000 × g. The supernatant was decanted, and the step with the sodium phosphate solution buffer was repeated twice. The soil pellet was re-suspended in 1 mL lysis solution I, briefly, 1 mL of lysis solution II, and 0.5 g of sterile sand (free of organic matter) were added and gentle mixing. Microbial lysis was done with Fast-Prep®-24 (MP Biomedicals™) at 5.5 m s-1 for 30 s. The suspended soil in both lysis solutions was passed through a cycle of freezing at -40 °C for 60 min, and thawing at 65 °C for 20 min. After freezing/thawing, samples were then centrifuged at 7700 × g for 10 min. The supernatant was transferred to a new sterile conical tube and mixed with 600 μL of EDTA solution and 300 μL potassium acetate solution, briefly, incubated at on ice for 30 min, and centrifuged for 10 min at 3000 × g. The supernatant was transferred to a new sterile conical tube and mixed with an equal volume of organic solvent mixture, centrifuged for 10 min at 3000 × g. The aqueous phase was recovered and washed with organic solvent mixture, and this step was repeated twice. The supernatant solution was transferred to a clean tube. Saline polyethylene glycol (PEG) in equal volume was added to the aqueous phase obtained, and incubated overnight at 4 ºC, and then centrifuged at 4 °C for 10 min at 12 000 × g. The supernatant was decanted. The DNA in the pellet was washed with 5 mL 70% cold ethanol and air-dried. The pellet of crude DNA extracts was re-suspended in 100 μL bi-distilled H2O. The DNA extracted was stored at -20 °C until used for PCR amplification.
Table 2: Composition of lysis solution and regents used during DNA extraction.
Solution |
Composition |
Sodium pyrophosphate solution |
0.15 M Na4P2O7 |
Sodium phosphate buffer solution |
0.12 M NaH2PO4, pH 8.0 |
Lysis solution I |
0.15 M NaCl, 0.1 M EDTA, pH 8.0 |
Lysis solution II |
0.1 M NaCl, 0.5 M Tris-HCl, pH 8.0, 12% SDS |
EDTA solution |
0.5 M EDTA, pH 8 |
Potassium acetate |
5 M CH3CO2K, pH 5 |
Saline 13% PEG |
13% PEG [8,000 MW], dissolved in 1.6 M NaCl |
Organic solvent mixture |
CHCl3:isoamyl alcohol (24:1) |
The Texcoco soils were pre-treated with Sorensen solution buffer PBS to prevent salt interferences and improve DNA extraction with the Mobio extraction kit. Soils were desalted by adding 10 mL of the Sorensen PB solution (pH 8.0), high speed vortexed for 1 min, let to settle for 10 min, and then centrifuged at 7700 × g at 15 °C for 10 min (Guo et al., 1997). Four samples of 0.25 g of pre-treated soil were extracted for DNA using the Mobio extraction kit following the manufacturer recommendations (Mo Bio Laboratories, Carlsbad, CA, USA) and then pooled, i.e., a total of 1.0 g of soil was extracted.
A positive control PCR was done using DNA extracted from Pseudomonas syringae pv. phaseolicola. The axenic strain P. syringae was extracted using the Fungal/Bacterial miniprep kit following the manufacturer’s protocol (Zymo research, Irvine, CA, USA).
PCR amplification of soil DNA
The DNA obtained was used as a template for bacterial 16S rDNA gene amplification by PCR using the universal primer 27F (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) (Yeates, Gillings, Davison, Altavilla, and Veal, 1997). Conditions were as follows: initial denaturation at 94 °C for 3 min followed by 35 cycles at 94 °C (1 min), 55 °C (1 min), and 68 °C (2 min) and ending at 68 °C (7.5 min). The reaction mixture contained the following: 1 μL template DNA (<20 ng μL-1) extracted with either the commercial kit or the developed method, with or without 6 mg mL-1 bovine serum albumin (BSA), 1X Pfx amplification buffer, 0.3 mM dNTP, 1.0 mM MgSO4, 0.3 mM primer mix, and 0.5 U Platinum Pfx DNA polymerase, Invitrogen.
Electrophoresis
The DNA extracted from soil and PCR-amplified DNA’s were gel electrophorized in TAE buffer (0.04 M Tris-acetate, 0.002 M EDTA [pH 8 ]) with 1.0% agarose (w v-1). Five μL extracted DNA and 3 μL PCR-amplified products were used during electrophoresis.
Determination of the purity and yield of DNA
The concentration of DNA in the extracts was determined spectrophotometrically as described by Sambrook, Fritsch, and Maniatis (1989) (Table 3). The purity of the DNA was determined by spectrophotometric readings at 230, 260, and 280 nm. The A260/280 and A260/230 ratios were calculated to evaluate levels of protein and humic acid impurities, respectively, in the extracts (Ogram Sayler, and Barkay, 1987; Steffan and Atlas, 1988).
Table 3: Crude DNA and ratios for different soil samples extracted using the Kit Mo BioTM and the improve technique in this study.
Soil |
Electrolytic conductivity |
Power Soil DNA kit |
Technique developed |
|||||
DNA |
A 260/280 |
A 260/230 |
DNA |
A 260/280 |
A 260/230 |
|||
dS m-1 |
µg g-1 of soil |
µg g-1 of soil |
||||||
Texcoco-1 |
39.9 |
2 |
1.47 |
0.45 |
3.67 |
1.18 |
1.05 |
|
Texcoco-2 |
9.8 |
6 |
1.41 |
0.55 |
2.35 |
1.17 |
1.06 |
|
Texcoco-3 |
2.6 |
7.75 |
1.41 |
1.09 |
3.67 |
1.38 |
0.95 |
|
Acolman |
0.8 |
12.75 |
1.64 |
0.48 |
4.5 |
1.09 |
0.83 |
|
Results and Discussion
Numerous methods have been developed to extract DNA from soil, but most of the proposed extraction methods are limited to certain environments and limit the extraction process to a defined group of soils (Alm and Stahl, 2000; Hurt et al., 2001; Zhou, Xia, Huang, Palumbo, and Tiedje, 2004; Devi et al., 2015; Guerra et al., 2020). Soil of the former lake Texcoco is alkaline-saline, allophanic with few aggregates and with a large organic matter content thus often containing large amounts of humic and fulvic acids. No DNA extraction technique has yet been described for this type of soil with sufficient yield and specific details in the extraction steps.
We extracted DNA from the former lake Texcoco soils and an arable one using an extraction technique based on the methods of Tsai and Olson (1991), Erb and Wagner-Döbler (1993), Guo et al. (1997) and Valenzuela-Encinas et al. (2008). In addition, the commercial extraction kit Power Soil DNA (Mo Bio™ Laboratories, Inc.) was used as a reference technique. The extraction of DNA from Acolman, Texcoco-2 and Texcoco-3 soils with the commercial kit was successful but failed for Texcoco-1 soil (Table 3, Figure 2).
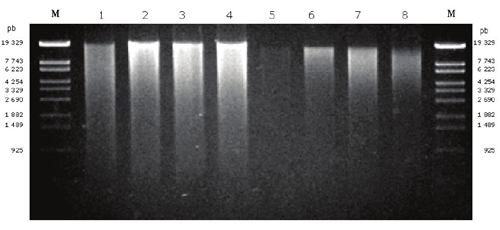
Figure 2 Total DNA from soils of Texcoco and Acolman. The agarose gel shows extracted DNAs. Lane 1, Texcoco-1; lane 2, Texcoco-2; lane 3, Texcoco-3; lane 4, Acolman extracted with the proposed method; Lane 5, Texcoco-1; lane 6, Texcoco-2; lane 7, Texcoco-3; lane 8, Acolman extracted with Power Soil DNA kit.
The low level of microbial biomass is the characteristic of alkaline soils and an efficient lysis procedure is required to obtain high yield and quality of DNA during the extraction. The use of a combination of methods, e.g. freezing-thawing process and mechanical disruption with Fast-Prep24, enhance the yield and quality in Texcoco-1, i.e. the soil with both higher EC and pH (Table 1). The efficiency of an extraction protocol could be measured comparing the time effort and reagents used in the process. Compared with DNA extraction method used in Texcoco soils (Valenzuela-Encinas et al., 2008) and saline-alkaline soil of Xochimilco, Mexico (Embarcadero-Jiménez et al., 2014), the proposed method employed minor use in reagent and time in the step of freezing-thawing process.
The PCR amplification was not possible using the DNA samples of the Texcoco and Acolman soils (Figure 3, line 2 to 9), i.e. no clean and/or bright bands of PCR products were obtained in samples without BSA. These might be related to impurities, such as protein, humic and fulvic acids (Table 3), so we recommend a purification step previous amplification to improve DNA quality. To avoid latter purification steps, Verma and Satyanarayana (2011) employed powdered activated charcoal and polyvinyl-polypyrrolidone to eliminate humic substances during soil DNA extraction. These reagents could by implement in further studies to improve the proposed extraction method.
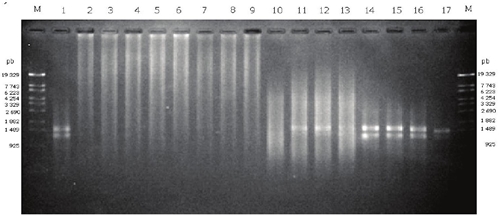
Figure 3: The agarose gel display PCR products. Lane 1, positive control using DNA of Pseudomonas syringae pv. phaseolicola; lane 2-9 without BSA. Lane 2, Texcoco-1; lane 3, Texcoco-2; lane 4, Texcoco-3; lane 5, Acolman extracted with Power Soil DNA kit; Lane 6, Texcoco-1; lane 7, Texcoco-2; lane 8, Texcoco-3; lane 9 Acolman extracted with the technique reported. lane 10-17 with BSA 6 mg mL-1. Lane 10, Texcoco-1; lane 11, Texcoco-2; lane 12, Texcoco-3; lane 13, Acolman extracted with Power Soil DNA kit; Lane 14, Texcoco-1; lane 15, Texcoco-2; lane 16, Texcoco-3; lane 17 Acolman extracted with technique developed.
Microbial growth in alkaline saline environments is exceedingly slow, as a proportionally high percentage of the energy generated is used for maintenance purposes, so the total soil microbial population is low (Oren, 1999). We, therefore, used 1 g of soil for DNA extraction technique described here. Ma et al. (2004) even used 10 g soil when extracting DNA from sediments of the Inner Mongolian Baer Soda Lake with similar characteristics as soil of Texcoco. Despite this, good extraction yields were obtained with the commercial kit and with the proposed method. However, differences were observed in the amplification process with BSA.
In our study, the inhibition of PCR by several impurities has been avoided by adding BSA (6 mg mL‑1) to the reaction buffer (Figure 3, line 11 to 17). BSA can eliminate a variety of contaminants which can bind and inactivate proteins or enzymes, and thus inactivate DNA polymerase. Bovine serum albumin has been used frequently during isolation of organelles and plants enzymes to sequester and eliminate endogenous phenolic contaminants, also BSA bind to lipids with hydrophobic forces and anions because of this high content of lysine (Loomis, 1974). Bovine serum albumin has been added to PCR buffer to reduce the activity of endogenous protease activity contained in some samples (Cho et al., 1996), so BSA may provide an alternative substrate and causing the DNA polymerase to remain protected from denaturing substances (Kreader, 1996). The extracted DNA with the proposed method was of good quality with few impurities after PCR amplification using BSA added to the reaction buffer. Additionally, few impurities were detected in the electrophoresis gel (Figure 3). The commercial kit allowed to amplify the DNA extracted from the soils of Acolman, Texcoco-2 and Texcoco-3, but not from the Texcoco-1 soil. The Texcoco-1 soil had a higher pH and EC than the other two Texcoco soils, which hampered the DNA extraction. In addition, the Texcoco-1 soil had a higher mineral salt and organic matter content which might have been coextracted with the DNA and could not be removed with the commercial kit. It is known that the presence of humic and fulvic substances inhibit the activity of polymerase enzyme (Tebbe and Vahjen, 1993). Additionally, organic compounds, such as bilirubin, bile salts, urobilinogens and polysaccharides, might also inhibit the PCR amplification (Wilson, 1997).
The concentration of BSA 6 mg mL-1 used in the amplification buffer was larger than the 3 mg mL-1 used by Romanowski, Lorenz, and Wackernagel (1993) and the 0.4 a 1.0 mg mL-1 used by Throbäck, Enwall, Jarvis, and Hallin (2004) for the amplification of DNA samples. Good results were obtained with the PCR amplifications; however, the 260/280 and 260/230 ratios were lower than the recommended values for pure DNA. The A260/230 ratio is indicative for contamination with humic acids and ratios >2 indicate pure DNA, while the A260/280 ratio is indicative for contamination with proteins and ratios >1.7 indicate pure DNA. For instance, a previous investigation showed that the extraction of DNA using two commercial kits, i.e. FastDNA® SPIN Kit for Soil and ZR Soil Microbe DNA Kit Miniprep™, from biochar-amended soil resulted in low values of purity, i.e. A260/230 <0.5 (Leite et al., 2014). In addition, Antony-Babu et al. (2013) reported A ratios >0.8 in DNA extracted using commercial kits from calcareous soils. Therefore, A260/230 ratios obtained using commercial kits are lower than those obtained using our proposed method. Pre-treatment of soil with phenol:chloroform-guanidine thiocynate or Triton X-100-skim milk during the DNA extraction has shown the improvement in DNA quality (Antony-Babu et al., 2013; Cheng et al., 2016). However, the implementation of any of those steps increase the time effort, reagents, and cost of the extraction technique.
Conclusions
In this study, we modified a method to extract DNA from alkaline-saline soil so that it could be used as a template for PCR amplification and sequencing. The proposed method for the extraction of DNA from alkaline saline allophanic soils with large organic matter contents adds important steps that reduce the time in the extraction procedure and a lesser use of reagents. Freeze and thaw cycles in combination with Fast-Prep-24™ equip showed good quality and yield in the Texcoco-1, soil with the highest pH and electrolytic conductivity. The use of BSA in PCR reaction buffer showed an improved DNA amplification in all soil samples obtained with the proposed method and three from four soils samples using the commercial kit.
The proposed method successfully extracted DNA from extreme saline-alkaline soils with large organic matter content that might be applicable for others soil types with extreme characteristics such as high pH and electrolytic conductivity values.
Data Availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.











 nueva página del texto (beta)
nueva página del texto (beta)