Introduction
Molecular oxygen has many different functions in the metabolism of microorganisms. It is the most important chemical agent that controls redox potential in aquatic ecosystems, and it is the main terminal electron receptor for aerobic microorganisms, both heterotrophic or chemolithotrophic. For this reason, oxygen concentration is one of the main environmental variables that modify microorganism distribution (Ferrara-Guerrero and Bianchi, 1990; Atlas and Bartha, 2006). Not only is molecular oxygen fundamental for the cell, but it is also the origin of multiple toxic reactions that induce cellular responses of diverse forms. Bacteria respond to the favorable oxygen concentrations with positive aerotaxis and to the unfavorable concentrations with negative aerotaxis. In many biotopes such as soils and sediments, aerobic bacteria constitute a considerable part of the microbial population (Garrity et al., 2004). Nevertheless, in stratified ecosystems there is a group of microaerobic bacteria that develop better at pO2 of 0.2 to 7% (Ferrara-Guerrero et al. , 1993).
In sediments, the concentration of available oxygen is low, allowing the formation of microoxic biotopes (i.e. with oxygen concentrations lower than 5%, approximately 10 mM O2 is dissolved). An example of microoxic habitats is that of nodules of leguminous roots with oxygen concentrations below 1 µM. Conditions of poor oxygenation promote the growth of diazotrophic symbiotic bacteria (e.g. Rhizobium or Bradyrhizobium ). Microaerobic bacteria require O2 for their growth. Nevertheless, high concentrations of this gas act as an inhibitor. On the other hand, terminal oxides of these bacteria have apparently low values of KM (affinity of an enzyme for its substrate) at O2 concentrations below 1 mM (Imhoff, 2006).
Microaerobic bacteria are part of a very diverse group of great physiological importance. They are very difficult to isolate, however, because of their critical oxygen demands. The group of microaerobic bacteria has a fundamental role in nitrogen recycling in aquatic ecosystems since many of them are involved in molecular nitrogen fixation (N2) (Reimers, 1989) and in organic matter mineralization, as in the case of some saprophytes described by Barrera-Escorcia and Namihira-Santillan (2004). Some of the genera that are important for farming are Azotobacter , Bradyrhizobium , and Rhizobium used intensively in agricultural activities since they contribute up to 50% of plant nitrogen needs (González et al., 1992; Dibut-Álvarez, 2009). Helicobacter pylori , the causal agent of peptic ulcers, is an example of microaerobic species of medical importance (Bergers, 2000). Little is known about the chemical characteristics of microaerobic bacteriobenthos cell membranes. Most of the information on this subject is found for aerobic and anaerobic groups from land environments (Buyer et al., 2002; Jafra et al., 2006) but not from sediments of aquatic environments. Specifically, the information on the fatty acid methyl esters (FAME) profiles of free-living microaerobic bacterial cell membranes is not very abundant. However, the FAME analysis of microaerobic bacteriobenthos cell membranes can be a powerful tool for detecting and differentiating bacterial species from sediments, as reported by Vainshtein et al. (1992) and Hippe et al. (2003) for some Desulfovibrio species, by Webster et al. (2006) for the genus Desulfococcus , and by Zhadanov et al. (2006) for Pseudomonas aurantiaca . However, careful standardization of growth conditions is needed because membrane composition is strongly affected by environmental conditions and by the nature of the culture medium (Buyer et al. , 2002). Certain microbial taxonomic groups can be identified by the presence of specific fatty acid biomarkers (e. g. isoprenoid quinones) and other molecules, which offer tools for studying the structure and changes in microbial community compositions as a result of environmental changes and the introduction of pollutants into aquatic systems (Thompson et al., 1993; Paige et al., 2002; Piotrowska and Mrozik, 2003; Munn, 2004; Mrozik et al. , 2005).
Most of the available information on FAME profiles corresponds to genera already described, such as Azotobacter, Bradyrhizobium, Rhizobium (with some facultative microaerobic species), Actinobacteria, Vibrio, Bacillus, Ureibacillus (strict aerobic), Clostridium (with strict and facultative anaerobic species) (Hamamoto et al., 1995; Graham et al., 1995; Dunfield et al., 1999; Gagné et al., 2001; Paige et al., 2002) due to their medical, ecological and economic importance, while only a few studies exist on the fatty acid (FA) profiles of some benthic bacterial genera (Gordon et al. , 2006). In this context, this investigation aimed to study the diversity of benthic bacteria populations with diazotrophic microaerophilic metabolism isolated from sediments of three aquatic environments, through their responses to different physiological, and biochemical tests, their molecular identification with 16S rDNA sequencing analysis and characterization of their FAME profiles.
Materials and methods
The sediment samples were collected in three mexican aquatic ecosystems: a) Lake Xochimilco, located in the SE of Mexico City (98° 57' and 99° 22' W, 19° 03' and 19° 36' N), at an altitude of 2500 m, is formed by canals which are currently fed by treated sewage water from the city, and its mud is used for cultivation in the intercanal areas known as 'chinampas' (Juárez et al. , 2003); b) Sontecomapan Lagoon, located in the southern part of the state of Veracruz (95° 00' and 95° 00' W, 18° 30', 18° 34' N), has an irregular shape and an area of 891 ha. In this shallow water body (2 m average depth), water is exchanged between the lagoon and the ocean through a natural mouth on the NW edge of the lagoon (Reséndez, 1982) and; c) an area in the southwestern part of the Gulf of Mexico (20° 12' and 21° 46' N, 92° 24' and 93° 24' W), in the San Pedro and Grijalva River plumes, with a salinity of 36 PSU (Suárez and Gasca, 1992; Salas de León et al., 2004).
Sediment Samples
Upper layer sediments (0-10 mm depth) from shallow sites (ten from Sontecomapan Lagoon and seven from canals of Lake Xochimilco 'chinampas' sites) were collected with an acrylic hand corer (50 mm i.d. and 260 mm long). Two milliliters of sediment were taken from the corer using a sterile syringe without the needle and preserved at -20 °C with 18 mL of a sterile 20% glycerol solution until processing in the laboratory (Ferrara-Guerrero et al., 2007).
Sediment samples from river plumes were collected with a Van Veen grab during cruises on the Oceanographic Ship Justo-Sierra (UNAM, Mexico). A 2 cm3 subsample of each grab launch was taken using a sterile syringe without the needle, avoiding disturbing the surface (Ferrara-Guerrero et al., 1993). Salinity of the interstitial water was measured with a refractometer ATTAGO(r) model 3T equipped with a temperature compensator. The interstitial water was obtained from the undisturbed sediment cores using a 5 mm outer diameter (o.d.), 10 cm long capillary tube with a series of millimetric perforations along the first centimeter of the capillary tube tip and the upper bound was connected to a hose that was attached to a 50 mL syringe.
Fifty three bacterial isolates from the upper layer sediments of Lake Xochimilco were obtained; 43 of them were able to grow in suboxic conditions. Thirty-seven bacterial isolates were obtained from the Sontecomapan Lagoon (18 grew optimally in suboxic conditions) and 37 from the Gulf of Mexico (of these, 17 were microaerobic).
Oxygen tolerance (pO2 of 0.4 and 21%) and molecular nitrogen fixing capacity tests were performed on the 78 isolates able to grow in suboxic conditions. In the follow-up of their response to this test, only 15 isolates (5 from each ecosystem) were selected since they grew at pO2 of 4%, had N2 fixing capacity and did not respire sulfates.
Cultivation and Purification of Microaerophilic Diazotrophic Bacteriobenthos
In this study, to capture a diverse array of microaerophilic bacteria able to fix N2, a semisolid mineral medium (2 g bacteriological agar L-1) was used, with 0.5 g L-1 calcium succinate (Sigma) as the only carbon source, no nitrogen source and pH 7.5 (Ferrara-Guerrero et al., 1993; Ferrara-Guerrero and Bianchi, 2000). For isolating microaerobic bacteria with capability of fixing N2, the method of serial decimal dilutions in 0.9% of NaCl solution was used. An aliquot of 1 mL of each dilution was inoculated into 9 mL of the semisolid growth medium, previously liquefied in a warm bath, and incubated at 20 °C for two weeks (Ferrara-Guerrero et al. , 1993). The microaerobic microbial growth was observed as subsurface rings of growth. Each growth ring was extracted with a sterile Pasteur pipette under a continuous N2 flow to prevent oxygen from entering the culture medium. It was then transferred to Hungate tubes, which were gasified with a mix of gases (4% O2 and 96% N2, v/v) (Praxair(r), standard calibration) and contained 9 mL of liquid mineral medium enriched with 0.1 ml L-1 of Balch's vitamin solution (Balch et al., 1979), and incubated at 25 ºC at 70 rpm for seven days. The mixed bacterial cultures were purified by series of decimal dilutions in mineral liquid medium gasified with 4% pO2. The purity of the bacterial culture was verified in plates incubated in suboxic atmosphere (manometer Bioxon(r) jars for anaerobic cultures), and by observation in a phase contrast microscope 100x (Olympus Bimax 50). Pure cultures were conserved in Hungate tubes containing 9 mL enriched liquid growth medium at pO2 4%.
Tolerance to Different Oxygen Concentrations
To determine their tolerance to oxygen, the microaerophilic isolates were incubated in jars for anaerobic cultures at different pO2 (21% O2 and 4% O2 balance 96% N2) and anoxic conditions (10% N2 balance 90% CO2) (PRAXAIR(r)).
Morphological and Physiological Tests
The characterization of microaerophilic isolates, incubated under low oxygen atmosphere (pO2 4%) in anaerobic culture jars, included four morphological (colonial form, cell form, mobility, coccoid bodies presence) and eight physiological (Merck, 2000)(Gram-stain response, capsule formation, cytochrome oxydase and catalase activities, nitrate and sulfate respiration, glucose oxidation and fermentation) tests (Stanier et al., 1966; Meynell and Meynell, 1979).
Methyl Esters Determination
FAME type and concentration of cell membranes of pure bacterial isolates were determined with the gaseous phase chromatography method (Badings and Joung, 1983; Findlay and Dobbs, 1993). For fatty acid (FA) extraction, five Petri dishes with triptone-yeast extract solid medium were incubated under low oxygen conditions (pO2 4%) to obtain massive cultures of each pure bacterial isolate (Dunfield et al., 1999). Bacterial biomass was then collected by washing with 3 to 4 mL of 5% formaldehyde solution and centrifuged at 6500 rpm for 15 min at 4 °C. The cell pellet was washed twice with a physiological solution (0.85% NaCl). The biomass obtained was deep frozen for 24 h at -70 °C and then lyophilized for 3 h at 7 kPa vacuum. Twenty milligrams dry weight were methylated with 0.1 mL sodium methoxide 0.5 N and 0.9 mL nanograde petrol (Díaz-González et al., 2002). A 1 µL aliquot was injected in a gas chromatograph (Perkin Elmer Autosystem 9000 Model) equipped with a flame ionization detector and a 100 m long smelt silica capillary column with syloxene and a high thermal resistance carbonate (SEG HTS) base, aluminum coated; helium (4.6) was used as a carrying gas. As a standard, a mix of 37 fatty acids and methyl acids (FAME) SUPELCO(r) (47885-U) was used (Pérez et al., 1997). The separation temperatures of the fatty acid methyl esters were 140 °C (5 min) to 240 °C at 4 °C min-1, hold 15 min.
Percentage of the concentration and relative frequency for each FA was calculated by dividing the number of times each FAME type appeared by the whole number ofisolates (15).
Extraction of DNA, and Amplification and Sequencing of the 16S rDNA
Extraction of the genomic DNA was accomplished from 20 mg of humid biomass using the commercial purification kit Fast DNA for Spin Kit Soil (Ros et al., 2006). Amplification of the 16S rDNA segment was carried out considering 50 ng of genomic DNA, using the universal primers rD1 (5´-AAGGAGGTGATCCAGCC- 3´) and f D1 (5´-AGAGTTTGATCCTGGCTCAG- 3´) (William et al., 1991). Amplification conditions were the following: 10 min at 95 °C; 35 cycles, each consisting of 1 min at 95 °C, 1 min at 45 °C, and 1 min at 72 °C; and finally 10 min at 72 °C (Laguerre et al., 1994; Puerta and Urueña, 2005). The amplified products (1500 bp) were visualized by means of electrophoresis in 1.2% agarose stained with ethidium bromide solution; the amplicon was purified with the QUIaquik (Quiagen(r)) package and sequenced. The results of sequencing were analyzed in DNA Star version 4.0(r) and DNA Man(r) programs.
Statistical Analysis
The results of 93 morphological, physiological and chemical tests performed on each of the bacterial isolates were submitted to Cluster Analysis with Euclidean distance as a similarity measure, using Ward's minimal variance method as an algorithm for group formation (Pielou, 1984) and Statistica(r) software (StatSoft Inc. Tulsa, OK, USA). To determine the contribution of each FA to the total variance of FA composition in the bacterial isolates, a principal components analysis, based on the diversity of methyl esters (37), was done. To define with precision the criteria for the FA classification according to its frequency and abundance, an Olmstead-Tukey graph was used, in which frequency and abundance were represented in a scatterplot, and the two axes were divided according to their corresponding median. The resulting quadrants correspond to dominant, frequent, rare and abundant FA. This approach has been successfully used for classification of plant and animal communities (Sokal and Rohlf, 1995).
The bacterial strains with microaerophilic metabolism used as references in all morphological, physiological and chemical tests were Azospirillum lipoferum DSM2292 (Tarrand et al., 1979) and Magnetospirillum magnetotacticum DSM3856 (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig Germany) (Maratea and Blakemore, 1981).
Results and discussion
Tolerance to Different Oxygen Concentrations
The isolates were capable of growing in oxic and suboxic conditions (pO2 of 21 and 4%). No isolate grew under strict anaerobic conditions (10% N2/90% CO2) (Table 1).
Morphological and Physiological Tests
All isolates were motile rods, 60% exhibited capsule and only 33.3% refractive bodies. Sixteen percent of the bacterial isolates were Gram-negative, and of these, 20% presented the cytochrome oxydase enzyme, and all were positive for catalase activity. Only three isolates from Lake Xochimilco did not respire nitrate; in contrast, none used sulfate as a terminal electron acceptor. Six isolates were able to oxidize glucose, and only one was unable to ferment it (Table 1).
Fatty Acids Identification
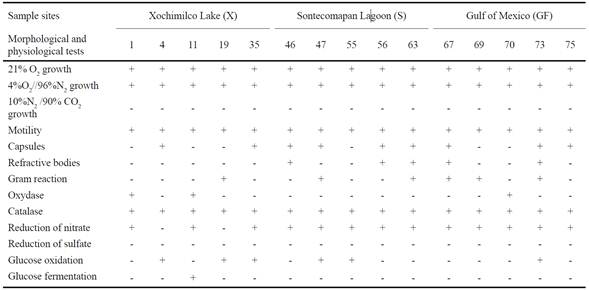
The FAME analysis performed on each of the 15 pure bacterial isolates proved the existence of 31 different types of FA in cell membranes (Figure 1). The highest variability was found in isolates from Sontecomapan Lagoon (26 types), followed by those from the Gulf of Mexico (22) and Lake Xochimilco (19).
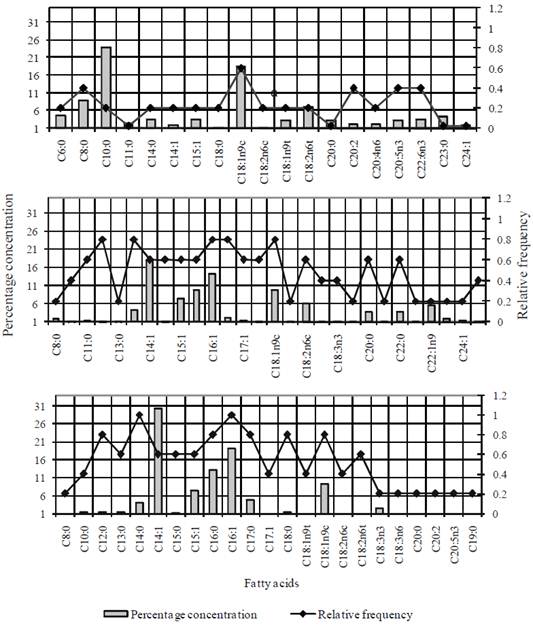
Figure 1 Relative frequency and concentration (% p/p) of the different methyl esters identified in the microaerophilic bacteria community isolated from superficial sediments of canals of Xochimilco Lake (a), Sontecomapan Lagoon (b) and Gulf of Mexico (c).
Relative frequency of FAME according to its percentage concentration (% w/w) was calculated. The most representative FAs from Lake Xochimilco were the C8:0, C18:1n9C, C20:2, C20:5n3 and C22:6n3 (Figure 1a), and for Sontecomapan Lagoon and Gulf of Mexico isolates were C11:0, C12:0, C13:0, C14:0, C16:0, C16:1, C17:0, C18:0 and C18:1n9c (Figures 1b and 1c). The FA which showed the highest concentration percentages were C10:0, C14:1, C16:0, C16:1 and C18:1n9c types, in the three environments (Table 2).
Table 2 Fatty acid (FA) profiles in the membranes of bacterial isolates from each study zone and proportional concentration in percentage.

The lowest percentages of FAME were found in isolates from the Gulf of Mexico. Only C14:1 showed a concentration higher than 30%, contrasting with the others which were mostly less than 10% (Figure 1c).
Some FAME types were characteristic of the bacterial isolates from a particular ecosystem. In this way, the C6:0, C20:4n6, C22:6n3 and C23:0 were identified as Lake Xochimilco isolates (X1, X4, X19 and X35), and C22:0, C22:1n9 and C24:0 as Sontecomapan Lagoon isolates (S47, S55 and S56), while no characteristic FAME were found in the isolates from the Gulf of Mexico. FAME diversity and percentage concentration, in the three study regions, are shown in Table 2.
Statistical Analysis
A cluster analysis was performed considering morphological and physiological variables as well as FAME profiles from the 15 bacterial isolates, including two reference strains (A. lipoferum and M. magnetotacticum ). At a Euclidean distance of 28, two phenons and three pairs (Figure 2) were identified. Pair 1 and phenon 1 gathered the majority of the isolates from Sontecomapan Lagoon sediments, where salinity fluctuated between 5 and 18 PSU. Phenon 1b gathered most of the isolates from Sontecomapan Lagoon and the Gulf of Mexico, where the salinity fluctuated between 14 and 36 PSU. Group (a) of phenon 1 clustered the strains GF73 (Bacillus sp.), GF70 (Pseudomona stutzeri ) and GF69 (Bacillus sp.). Even though strains GF69 and GF73 are Gram positive and do not produce oxidase, strain GF69 is closely linked to P. stuzeri (GF70) because they do not oxidize glucose, there is no capsule. In addition to the fact that these three strains equally present, the same five AG in their cell membrane (Table 2). Phenon 2 was integrated by bacterial isolates from Lake Xochimilco sediments, which is a freshwater ecosystem. Furthermore, pairs 1 and 3 gathered isolates able to ferment/or oxidize glucose, thus differing from phenons 1 and 2 of which only some of them are capable of doing so.
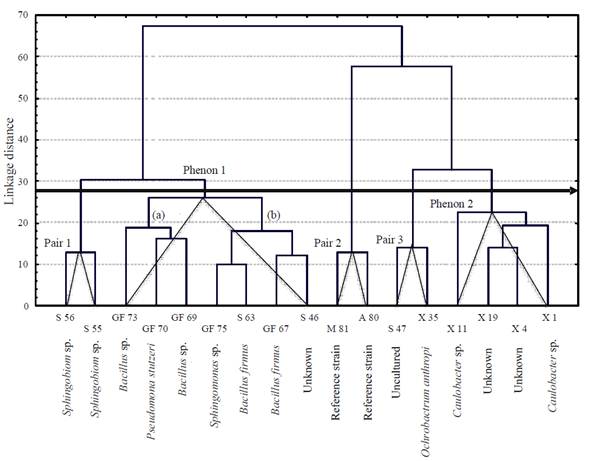
Figure 2 Dendrogram elaborated considering the different morphological, physiological and fatty acids tests done on the 15 bacterial isolates from the three study sites. Xochimilco Lake (X), Sontecomapan Lagoon (S), Gulf of Mexico (GF), and collection strains: Azospirillum lipoferum (A80, DSM2292) and Magnetospirillum magnetotacticum (M81,DSM3856).
The principal components analysis (PCA) based on FAME diversity provided 4 groups. Group 2 included the highest number ofisolates (9), all obtained from Gulf of Mexico and Sontecomapan Lagoon (Figure 3); this group showed highest FAME diversity (23 types), in which the most representative types were C12:0, C16:1, C17:0 and C18:1n9c. Groups 1 and 3 presented the lowest diversity with 8 and 9 FA types, respectively. Group 1 gathered isolates with FAME considered characteristic (C6:0, C23:0 and C22:0) ofisolates X1, X35 and S47; the presence or absence of one of these FAs is the feature that separates one isolate from the other and from the rest of the isolates of the group. A similar pattern happened in group 4, which presented the FAs C20:4n6 and C22:6n3.
Figure 3 Score plot of principal component analysis based on diversity of the fatty acids methyl esters present in cell membranes of 15 bacterial isolates from Xochimilco Lake (X), Sontecomapan Lagoon (S) and Gulf of Mexico (GF) sediments; bacterial isolates with characteristic fatty acids are shown within a doted circle.
The reference strains (A. lipoferum and M. magnetotacticum ) and isolate X11, all characterized by the FAs C20:0, C18:1n9t, C18:1n9c and C15:1, clustered in group 3. In general, the four axes explained 78.3% of the cumulative percentage of variance. Eigenvector 1 represented 57.3%, eigenvector 2, 24.9%, eigenvector 3, 53.4% and eigenvector 4, 68.7%.
According to Olmstead-Tukey plot, in which FA ofisolates from each study site are classified by categories (abundant, dominant, rare and frequent), most of the FA reported as rare were found in isolates from Lake Xochimilco and the Gulf of Mexico (Figures 4 and 6). This was expected; otherwise, most of FAME in isolates from Sontecomapan Lagoon was reported as frequent (Figure 5).
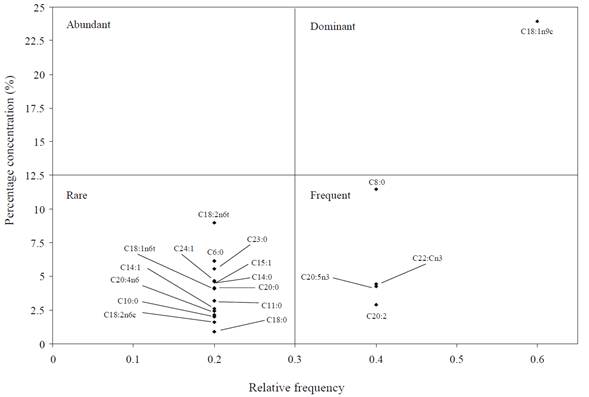
Figure 4 Tukey' plot in which categorization of fatty acids present in isolates from Xochimilco Lake is shown.
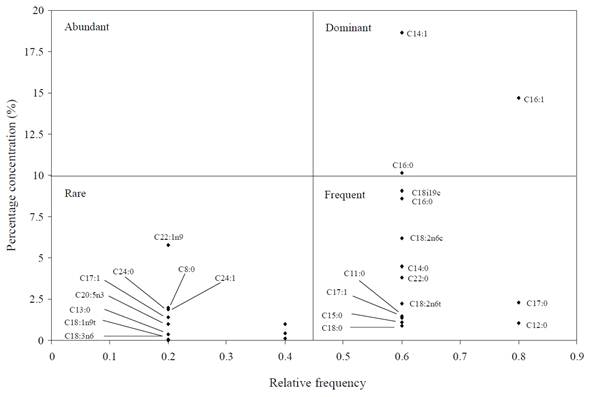
Figure 5 Tukey' plot in which categorization of fatty acids present in isolates from Sontecomapan Lagoon is shown.
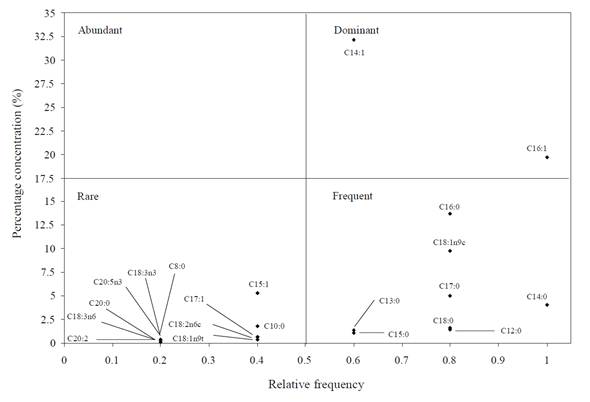
Figure 6 Tukey diagram in which categorization of fatty acids present in isolates from Gulf of Mexico is shown.
No fatty acid of the bacterial isolates from the three areas studied was placed in the category of abundant, and four of them were classified as dominant (C18:1n9c, C14:1, C16:0, C16:1); of these, the isolates from the Gulf of Mexico and the Sontecomapan Lagoon shared types C14:1 and C16:1.
Amplification and Sequencing of 16S rDNA
Sequences of 16S rDNA showed that the whole isolates presented high levels of similarity (98 to 100%) to eight sequences reported in the National Center for Biotechnology Information (NCBI), which correspond to eight bacterial species: Caulobacter sp., Ochrobactrum anthropi, Sphingobium sp., Bacillus firmus, Bacillus sp., Pseudomona stutzeri, and Sphingomonas sp. The sequences of the isolates X4, X19 and S46 did not match any sequence described in the NCBI. Isolate S47, which so far has been reported as uncultured, may correspond to the genus Ochrobactrum since in the dendrogram it was gathered at the same Euclidean distance (14) as O. anthropic , and in the NCBI records it had a similarity of 89% with this genus (Table 3).
Table 3 Identification of bacterial isolates following sequencing of 16S rDNA, and FA that were unique in some of the species identified.

All the strains exhibited facultative microaerobic metabolism since they grew in aerobic and microaerobic conditions and were not able to grow in strict anaerobic conditions. Most of them showed presence of cytochrome oxydase enzyme, and thus, none are able to survive under oxygen saturation conditions. Generally, microaerophilic microorganisms are well adapted to suboxic sediment microhabitats (Munn, 2004).
According to the obtained results, it is difficult to establish the composition of the native microbial communities of the suboxic sediments since their bacterial populations are not formed by a simple group; rather they belong to numerous genera, many of which are characteristic of soil ecosystems. In marine ecosystems, the most common bacterial metabolism is facultative anaerobic chemolithotrophic because the O2 gradient in these systems is more variable than in deep waters and sediments (Acuña et al., 2011).
Taxonomic studies in different aquatic ecosystems (lakes, lagoons, rivers and oceans) indicate that the most common bacteria in these environments belong to Pseudomonas , Aeromonas , Vibrio , Bacillus , Caulobacter , Acetobacter , Rhodobacter , Microccocus , Flavobacterium , Planococcus , Spirillum , Alcaligenes , Azospirillum , Aquaspirillum, and Magnetospirillum ; the last two are the most frequently reported genera (Miravet et al., 2003; Lin et al., 2011).
However, in this investigation, only the genera Pseudomonas (isolated from the Gulf of Mexico sediments), Bacillus (isolated from Sontecomapan Lagoon and Gulf of Mexico sediments) and Caulobacter (from Xochimilco channel sediments) were found; these gathered in different phenotypes formed clusters (Figure 2) according to their marine or fresh water origin. These results suggest that salinity is a determining factor in the physiological diversity of the microaerobic bacterial community.
The origin ofisolates S63 (Bacillus firmus ) and S46 (unknown) is in areas near the mouth of the Sontecomapan Lagoon, where salinity ranges between 30 and 40 PSU, and the connection with marine water is permanent; possibly for this reason, these isolates grouped in the same phenon separate from those from the Gulf of Mexico (GF75 Sphingomonas sp. and GF67 Bacillus firmus ).
Although pairs 1 (S56 and S55 Sphingobium sp.) and 2 (A. lipoferum and M. magnetotacticum ) gathered at the same Euclidean distance of 12 because they had similar morphological and physiological characteristics, they formed an independent cluster due to their FAME diversity. Pair 1 comprised 28 types and the collection strains had only 19. The most representative FAME in pair 1 were types C12:0, C14:0, C14:1 and C15:0, while for the collection strains the most representative were types C12:0, C18:0 and C18:1n9c.
The FA analysis has been widely used for taxonomic classification and bacterial phylogeny, as reported by Piotrowska and Mrozik (2003), who stated that these molecules could be used as powerful taxonomic tools. Zhadanov et al. (2006) described important differences in FA of Pseudomonas aurantiaca , which mainly correspond to types C16:0, C16:1 and C18:1, identified in Pseudomonas stutzeri (GF70) isolates from the Gulf of Mexico (Table 2).
Our results are consistent with those obtained by Frolova et al. (2005), Fang et al. (2006) and Gordon et al. (2006), who reported the FAs C13:1, C14:0, C15:0, C15:1, C16:0, C16:1 and C17:0 as characteristic of microorganisms and proteobacteria from marine sediments. Furthermore, some of the identified species had unique FA.
Although none of the 16S rDNA segment sequences ofisolates X4 and S46 matched any of those reported by NCBI, some of their cell membrane fatty acids do match those reported by Blazina et al. (2010) as characteristic of the genera Vibrio, Halomonas and Pseudoalteromonas (Table 3).
In the principal components analysis, group formation was determined by their FAME diversity; this confirms that FAME composition can differentiate a marked group ofisolates, as is the case of the isolate S47, which belongs to group 1 with only 4 types of characteristic FAME of bacteria isolated from Sontecomapan Lagoon sediments. This could explain the fact that this isolate was plotted far from the origin, marking it as distinctively different from the other isolates.
On the other hand, the strains utilized as references (A. lipoferum and M. magnetotacticum ) gathered in group 3 together with isolate X11 (Caulobacter sp.). Even though they belong to different genera, they share the presence of the FA C20:0, C18:1n9t and C15:1 in their cell membranes. The reason could be that these fatty acids are not markers of specific genera.
Most of the isolates of marine origin gather in group 2, in which FAME has the largest variation. Group 3 gathers the isolates from the Xochimilco freshwater system (Figure 3).
The Olmstead-Tukey analysis shows that the FAME characteristics of each environment are found in the category of rare (Figures 4, 5, and 6); particularly, these FAME types have a relationship to some strict microaerophilic benthic bacterial genera (S63 and GF73).
In contrast, it was observed that the Olmstead-Tukey diagrams from the Sontecomapan Lagoon and the Gulf of Mexico allowed the distinction of FAME according to the above mentioned categories, and some subgroups could even be distinguished within quadrants. These results suggest that FAME profiles of microaerophilic bacteria can be better understood by plotting them in this type of graph, which means that this chemotaxonomic approach is a precise tool for characterizing benthic bacterial communities and their relationship with their environment.
Conclusions
Even when the genera identified have been reported to date as strict aerobic or facultative anaerobic, the collection of strains exhibited facultative microaerobic metabolism since 100% grew well in aerobic and suboxic conditions (pO2 4%) and did not grow in strict anaerobic conditions.
Characteristic FAME that can be considered good chemotaxonomic tools and may be used as biomarkers of facultative microaerophilic bacteria presence in sediments from freshwater are C20:4n6, C22:6n3 and C23:0, and for brackish water C22:0, C22:1n9 and C24:0. Because culturing this type of bacteria is extremely difficult, FAME analysis can increase the opportunity to study bacterial diversity in a wide variety of ecosystems, even if uncultivable forms are present.
In this research, profiles of cell membrane fatty acids of Ochrobactrum anthropi (C8:0, C11:0, C18:2n6t and C23:0) and Sphingobium sp. (C22:0) that had not been previously reported have been established.
In spite of clear Gram negative and Gram positive chemical differentiation in the components of the microorganism cell membrane, it was clearly observed that C12:0, C14:0, C16:0, C16:1, C17:0, C18:0, C18:1n9c and C18:1n9t (lauric, myristic, palmitic, palmitoleic, haptadecanoic, stearic, oleic and eleadic acid methyl esters) can be found in both kinds of membranes.
In this study we observed that the diversity of microaerobic benthic bacteria was influenced principally by salinity, thus Bacillus and Sphingobium genera prevailed in brackish sediments and Caulobacter in freshwater sediments.











 nueva página del texto (beta)
nueva página del texto (beta)