Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de micología
versión impresa ISSN 0187-3180
Rev. Mex. Mic vol.34 Xalapa dic. 2011
Contribuciones
Molecular identification of the fungus causing postharvest rot in jackfruit
Identificación molecular del hongo causante de la pudrición postcosecha de la jaca
Juan Arturo Ragazzo–Sánchez1, Alika Gutiérrez–Escatel1, Guadalupe Luna–Solano2, Juan Florencio Gómez–Leyva3, Montserrat Calderón–Santoyo1
1 Laboratorio Integral de Investigación en Alimentos, Instituto Tecnológico de Tepic, Av. Tecnológico 2595, Tepic, Nayarit, México 63175.
2 Instituto Tecnológico de Orizaba.Av. Oriente 9 852, Orizaba, Veracruz. 94300 México.
3 Instituto Tecnológico de Tlajomulco, Carretera a San Miguel Cuyutlan km 10, A.P. 12, Tlajomulco de Zuñiga, Jalisco, México 45640.
Autor para correspondencia:
Montserrat Calderon–Santoyo montserratcalder@gmail.com.
Recibido 17 de agosto 2010;
Aceptado 7 de julio 2011.
Resumen
La jaca (Artocarpus heterophyllus) perteneciente a la familia Moraceae, es un fruto tropical originario de India y Malasia, actualmente cultivado en San Blas, Nayarit, México. La jaca tiene vida postcosecha corta (2–3 días después del corte), en su mayoría debido al ataque de hongos fitopatógenos, que todavía no han sido identificados para la jaca de Nayarit. El objetivo de este trabajo fue identificar los hongos que causan enfermedades postcosecha en jaca para desarrollar en un futuro nuevas prácticas postcosecha. Cinco hongos presuntivamente adscritos a los géneros Aspergillus y Penicillium fueron aislados de jaca con síntomas de enfermedad. La prueba de patogenicidad fue positiva sólo para Aspergillus sp. el cuál fue identificado por PCR como Aspergillus niger ATCC16888.
Palabras clave: hongos fitopatógenos, Artocarpus heterophyllus, Aspergillus niger, Penicillium.
Abstract
Jackfruit (Artocarpus heterophyllus) belonging to the family Moraceae, is a popular tropical fruit originated from India and Malasia and is now widely grown in San Blas, Nayarit, Mexico. Jackfruit has a short shelf life (2–3 days after harvest), mostly because of losses due to postharvest diseases caused by pathogenic fungi. These fungi have not been identified in jackfruit from Nayarit. The aim of this research is to identify the fungi that cause diseases in jackfruit in order to develop new post–harvest practices for the future. Five fungi, corresponding to the genus Aspergillus and Penicillium were isolated from deteriorated jackfruit. The pathogenicity test was positive only for Aspergillus sp. which after PCR analysis was identified as Aspergillus niger ATCC16888.
Key words: phytopathogenic fungi, Artocarpus heterophyllus, Aspergillus niger, Penicillium.
Introduction
Nayarit has integrated the jackfruit (Artocarpus heterophyllus) culture as an alternative crop due to its high demand on a regional, national and international level. Jackfruit is relatively new in our continent and its international interest has recently increased, especially because this fruit has unique organoleptic qualities (smell and taste) and nutritive properties (Ong et al., 2006; Saxena et al., 2008) that makes it desirable for commercial success in many countries, especially in the USA (SAGARPA, 2008). However, there are several factors negatively affecting the quality of this product, resulting in great losses. Fruit and vegetable decay rapidly, especially in tropical areas, due to microbial agents. This is mainly because of high temperatures and relative humidity, conditions highly encountered in Nayarit area. These conditions lend themselves to the attack of fruit by fungi, causing important produce losses. Generally, there is very little research available on jackfruit concerning postharvest diseases. In the past, these diseases have been studied in Asian countries. The fungi causing these diseases has been isolated and identified as Colletotrichum gloeosporioides Penz for jackfruit from Bangladesh (Basak, 1995) and Rhizopus artocarpi for jackfruit from India (Roy, 1983). In addition, a die–back disease caused by the bacteria Erwinia nigrifluens has been reported (Ton et al., 1990). No study has been conducted in order to establish the causal agent of postharvest diseases in jackfruit from Nayarit. Traditional practices for studies of fungi include conventional cultivation and microscopic identification (Deak, 1994). Identification of fungi species is based on mycelia (color, size and shape) and morphological characteristics (morphology, conidia size and morphology conidiophore) (Pitt and Hocking, 1999). These techniques require skilled taxonomists. Minor differences in medium composition can impair effective comparison of mycelia characters (Larone, 2002). Molecular techniques have been demonstrated as an effective and easy way to identify fungi. DNA–based assays are reliable to detect a variety of fungi. Specific primers have been selected to reliably amplify and analyze fungal DNA from culture. The polymerase chain reaction (PCR) has been used to selectively amplify DNA sequences of Basidiomycetes fungi (Jellison and Jasalavich, 2000). Some examples include the use of PCR and multiplex DNA amplification for Tuber species (Amicucci et al., 2000) or for Neofabraea in pome fruits (Gariépy et al., 2003), the use of PCR–based diagnostic using internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA) for Rhizoctonia species in rice (Johanson et al., 1998). Some specific marker genes based in laccase introns have been developed for multiplex PCR identification of Sclerotiniaceae (Hirschhäuser y Fröhlich, 2007). The aim of this work was to identify the agent causal of black rot fruit disease in jackfruit from Nayarit in postharvest by PCR techniques.
Materials and methods
Sample source
Jackfruits being in mature postharvest stage of fruit development and apparently uninfected were collected from Ixtapa de la Concepción, San Blas, Nayarit, Mexico. Fruits were transported to the Instituto Tecnologico de Tepic and stored in plastic boxes maintained semi–opened for avoiding anaerobic conditions by the CO2 production by the jackfruit. Boxes were installed in temperature–controlled chambers at 28 °C and relative humidity of 80 ± 2%. Changes were recorded every 24 h until disease visualization.
Fungi isolation and purification
Fruit epicarpical tissue segments of 1 cm2 were taken (half from affected and half from unaffected area) from 3 different zones: peduncular zone (I), equatorial zone (II) and lower zone (III). These samples were disinfected by immersion in 1% sodium hipoclorite solution during one min, rinsed in sterile water and transferred to potato dextrose agar (PDA) plates (acidified with 10% tartaric acid). Finally they were incubated at 28 °C during 48–72 h until mycelium development (NOM–111–SSA1–1994). Fungi mycelium were suspended in isotonic physiological solution (0.85% NaCl) and re–inoculated in PDA Petri dishes until purification.
Fungi pre–identification
The pre–identification of fungi isolated was performed using traditional morphological methods. Macroscopic (mycelium type, color and growth type) as well as microscopic (mycelium type, conidiophores morphology and spore morphology) characteristics were considered for identification (Larone, 2002).
Pathogenicity test
In order to discard secondary pathogens or saprophytic fungi from primary pathogens, a pathogenicity test based on Koch Postulates was applied. It consisted of isolation of the pathogen from the fruit presenting a typical disease, pathogen inoculation on a healthy fruit, disease development, reisolation of the pathogen and finally a comparison of first pathogens isolated with second one's as well as a comparison of symptoms first obtained with second symptoms. When symptom and pathogen characteristics were identical, the pathogenicity test was considered positive.
Spore suspension was prepared using one week old fungi cultures in PDA, 10 mL of a sterile physiologic solution (NaCl 0.9%) were added to the cultures and Petri dishes were moved gently. The liquid was filtered in sterile gauze and recovered in a dilution flask. Spore concentration was determined by microscopic counting in Neubauer cameras and spore suspension was diluted to reach a concentration of 105 spores mL–1 (Guiraud, 1998; Capdeville et al., 2007).
The surface of healthy fruits was disinfected with NaClO (20 (µg L–1) during 5 minutes and then rinsed with sterile water. Fruits were dried in a laminar flow chamber. Five fruits were inoculated by hyssop (a cotton swab was soaked in the spores suspension and placed on the epicarpial) and five by wound (fruits were wounded using a sterile scalpel and 150 (µL of a 105 spores mL–1 suspension were placed on the wound) for every isolated pathogen. Jackfruits were stored at 28°C in a controlled chamber with the same conditions mentioned above, until symptoms development. The new symptoms were compared with the previous symptoms (Albornett et al., 1994).
PCR identification
In order to confirm the genus and identify the specie for the fungus resulting positive in Koch test, identification based on the polymerase chain reaction, posterior Escherichia coli transformation and sequencing was applied.
DNA extraction was performed according to the protocol proposed by Lee and Taylor (1990). Fungi were grown in 100 ml Czapek broth at 28 °C for 48 h and centrifuged at 20000 g for 10 min. The precipitated was triturated in liquid nitrogen. 400 (µL lysis buffer (50 mM pH 7.2 HCl–Tris buffer, 50 mM EDTA, 3% SDS and 2% 2–mercaptoethanol) were added to 1.5 mL of triturated mycelium and incubated at 65°C for 1 h. Then, 400 µL of a solution chloroformphenol 1:1 were added. The mixture was centrifuged for 15 min at 14000 g until the supernatant was clear; 300–350 µL of the watery portion was removed and 10 µL 3M NaOAc as well as 0.54 isopropanol volumes were added. The mixture was turned gently and centrifuged for 2 min at 14000 g. The supernatant was discarded and DNA was washed with 70% ethanol. Tubes were dried under vacuum at 50 °C for 15 min. DNA pellet was dissolved in TE buffer (10mM Tris–HCl pH 8.0, 1mM EDTA; 15 µL). The final DNA concentration was between 0.1 to 10 g–1.
PCR was performed using the primers ITS1 and ITS4 (Atkins and Clark, 2004), DNA from Aspergillus sp. was used as a positive control and the negative control contained only the reactive. For amplification 5 µL of isolated DNA was used in 25 µL of PCR mixture. The reaction was performed in 10 mM Tris–HCl (pH 8.3), 2 mM MgCl2, 0.2 mM dNTP's, 20 nM of each primer and 1.5 U of Taq DNA polymerase. Amplifications were carried out in Genius Technethermocycler with thermal cycling parameters: initial denaturation at 94 °C for 2 min, followed by 30 cycles of heat denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min and final extension at 72 °C for 5 min. PCR products from these amplifications were separated by electrophoresis on 1.2% (w/v) agarose gel, stained in ethidium bromide and visualized using a UV transilluminator (Johanson et al., 1998).
Transformation of E. coli cells was performed according to Sambrook and Russell (2001) method using the 3954 pb TOPOVector (Carlsbad, CA, USA). 2 µL of PCR products were used in a 6 µL of transformation mixture containing 1 µL of TOPOVector and 10 mM NaCl. Tubes were incubated on ice for 5 min, followed by a heat shocking for 30 sec at 42 °C. Tubes were placed immediately on ice and 250 µL of SOC medium were added. LB agar Petri dishes containing 50 g ml–1ampicilin were inoculated with the mixture above and incubated at 37 °C for 8 h. White colonies were selected, reinoculated in ampicilin LB agar and incubated again at 37 °C for 8 h. White colonies were inoculated on ampicilin LB broth and incubated until turbidity. Finally, the 3954 pb plasmid was isolated according to the protocol proposed by Sambrook and Russell (2001). 1.5µL were transferred to microtubes and centrifuged at 10000 g for 5 minutes at 4 °C. The pellet was resuspended in ice–cold 100l 25 mM Tris–HCl–10mM EDTA pH 8.0. 15 mL 0.2N NaOH–1.0% SDS were added to the suspension and mixed gently. 15 mL ice–cold 7.5 M Ammonium acetate were added to the lysate and mixed by repeated gentle inversion. Lysate was centrifuged at 15500 g for 30 minutes at 4 °C. The resulting supernatant was transferred to new microtubes. An equal volume of phenol:chloroform (50:50) was added and mixed thoroughly by repeated inversion. The solution was centrifuged at 15000 g for 2 min at 4 °C. The resulting aqueous phase was recovered and two volumes of room temperature ethanol were added in order to precipitate nucleic acids. The mixture was left to repose 2 min and centrifuged at 15000 g for 5 min at 4 °C. Supernatant was discarded and 1ml of 70% ethanol was added to the pellet. DNA was recovered by centrifugation at 15000 × g for 2 min at 4 °C. Supernatant was discarded and 100l TE solution as well as 5 lRNAasa was added in order to eliminate RNA. Microtubes were incubated at 65 °C for 1 h. The mixture was washed with 100 µL phenol:chloroform (50:50) and precipitate was obtained by addition of 10 µL NaOAc and 100 µL isopropanol. DNA was washed with 70% ethanol, dried and resuspended in 50 µLL TE solution. PCR was performed under the condition enounced above and PCR products were separated by electrophoresis on 1.2% (w/v) agarose gel, in order to verify insert acquisition.
Ribosomal DNA was sequenced in the Sequencing Laboratory at CINVESTAV in Irapuato, Mexico. The sequencing machine ABI Prism 377 sequencer (Perkin Elmer Applied Biosystem, CT) was used. Nucleotides sequences w e r e c o m p a r e d a t B L A S T v 2 . 0 , (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi/) in order to identify the agent causal of postharvest disease of jackfruit.
Results
Isolation and pathogenicity tests
During the storage of jackfruit in high relative humidity conditions, disease symptoms appeared four to six days after incubation. Symptoms appeared principally as a mass of black or brownish spores, but mycelia growth presenting white, mustard yellow and green color was also obtained. After isolation from the damaged tissue, we obtained five species of fungi well differentiated by their micro and macro characteristics. Fungi were micro–cultivated for as many as 3 days to observe conidia and conidiophore morphology. Czapek agar was used for ensuring spores formation by osmotic stress. Four Aspergillus species and one for Penicillium were identified based on macro and microscopic characteristics found.
During pathogenicity tests, only one fungus produced disease symptoms with identical characteristics as the first symptoms obtained. Symptoms appeared as brown mycelium growth followed by abundant brown conidia development (Figure 1a). From these symptoms two fungi were isolated. One of this fungus presented different macroscopic characteristics from the five fungi first isolated, suggesting a contamination. This fungus was preliminary identified as Botryosphaeria sp. The second fungus presented macro and microscopic characteristics similar to those of the first isolated and inoculated fungus. Thus, pathogenicity showed to be positive for only one strain of Aspergillus sp. The observed characteristics for this specie of Aspergillus during the PDA culture were at first white, then turning to black, mostly consisting of a dense felt of erect conidiophores (Figure 1b). When observed at optical microscope (40), hyphae were septate and conidial heads were black and radiate. Conidiophores presented smooth–walled and hyaline stipes. The vesicles were completely covered with phialides. The phialides produced dense chains of round black conidias and were supossed to borne on metulae (Figure 1c). The remaining isolates were considered saprophytic fungi or secondary pathogens for jackfruit because they did not cause disease symptoms.
PCR identification
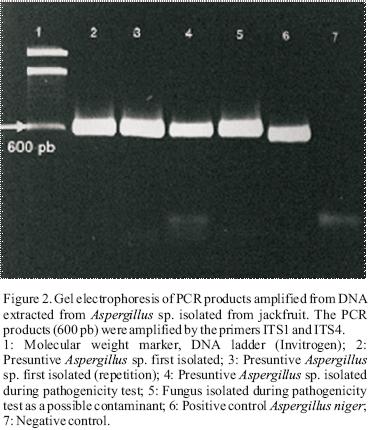
The identification of presuntive Aspergillus sp. was performed by PCR using the primers ITS1 (5'–CAACTCCCAAACCCCTGTGA–3') and ITS4 (5'–GCGACGATTACCAGTAACGA–3') (Abd–Elsalam et al.,2003). These oligos were obtained from the region ITS (Internal Transcriber Spacer) found in DNA ribosomal. The PCR products were visualized by electrophoresis and a 600 bp fragment was obtained (Figure 2).
After the transformation of Escherichia coli strains, a 1.2% agarose gel electrophoresis was used in order to confirm the acquisition of the insert (Figure 3). It was not possible to observe the insert in lanes 2 and 11 indicating a possible acquisition of the plasmid by E. coli but a lack of the insert in that plasmid.

The sequences obtained from the cloned E. coli strains were compared to those reported at "Basic Local Alignment Search Tool" (BLAST v2.0, http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi/) (National Center for the Biotecnology Information, NCBI, USA). This analysis shows that the resulting fungus pathogen for jackfruit was identified as Aspergillus niger strain ATCC16888 with a homology of 100% (Table 1).
The contaminant fungus was identified as Botryosphaeria rhodina with a homology of 100% (results not showed). Even if B. rhodina is recognized as a contaminant in fruits and vegetables, their macro and microscopic characteristics were not concordant with any fungus first isolated. Thus, it was concluded that B. rhodina was only a saprophytic fungus but not a pathogen of jackfruit.
Discussion
Aspergillus niger strains have been reported before as pathogens of fruit crops. Aspergillus rot is one of the most damaging postharvest diseases in apples during long term storage. Symptoms appear as brown areas with softening of flesh tissue with watercore (Kumpoun et al., 2003). A. niger is recognized by far as the most common species of Aspergillus present on grapes. It is responsible for the Aspergillus rot, but the major problem is their contribution to the ochratoxin A production. This toxin is produced primarily when Aspergillus infects berries before harvest (Hocking et al., 2007). Other Aspergillus species are recognized as pathogens in fruits because their capacity of producing mycotoxins: A. flavus and A. parasiticus in dates produce aflatoxins (Shenasi et al., 2002). A. westerdijkiae in orange fruit produce ochratoxin A (Marino et al., 2009).
Other pathogens have been identified in jackfruit from asiatic countries (Colletotrichum gloeosporioides Penz for jackfruit from Bangladesh and Rhizopus artocarpi for jackfruit from India, species of Aspergillus had not yet been reported as postharvest pathogens in jackfruit. Thus the A. niger strain ATCC16888 is possibly an endemic postharvest pathogen to jackfruit from the Nayarit area.
A. niger as well as A. fumigatus is considered an extremely dangerous pathogen. It causes aspergillosis in humans. A. niger is recognized as a producer of ochratoxin A (Hocking et al., 2007). Although most of these organisms cause severe illness in immune compromised individuals, sometimes otherwise healthy people may also become infected. These illnesses are common among people who work in the farming industry, and they are considered an occupational hazard. In addition, one of the diseases they cause is invasive pulmonary aspergillosis, which is difficult to diagnose (Larone, 2002). It is necessary to evaluate the possible production of mycotoxins by this fungus to determine a potential danger and to establish its epidemiology in order to develop adequate methods of control for this pathogen. More attention is necessary in order to control Aspergillus niger development in jackfruits.
Acknowledgments
A. Gutierrez–Escatel is indebted to COSNET (Mexico) for a research grant.
References
Abd–Elsalam, K.A.A., M.A. Abdel–Satar, M.S. Khalil, J.A. Verreet, 2003. PCR identification of Fusarium genus based on nuclear ribosomal–DNA sequence data. African Journal of Biotechnology 2(4):82–85. [ Links ]
Albornett, N., J. Yajaira, N.H. Sanabria de Albarracin, 1994. Diagnóstico de las enfermedades fingicas en frutos de lechosa (Carica papaya) y melón (Cucumis melo) para exportación, Revista Facultad Agronomia (Maracay) 20:13–20. [ Links ]
Amicucci, A., C. Guidi, A. Zambonelli, L. Potenza, V. Sstocchi, 2000. Multiplex PCR for the identification of white Tuber species. FEMS Microbiology Letters 189(2): 265–269. [ Links ]
Atkins, S.D., I.M. Clark, 2004. Fungal molecular diagnosis: a mini review. Journal of Applied Genetic 45: 3–15. [ Links ]
Basak, A.B., 1995. Fruit rot disease of jackfruit caused by Colletotrichum gloeosporioides Penz in Chittagong. Bangladesh Journal of Botany. 24(2): 197–199. [ Links ]
Capdeville, G., M. Teixeira–Souza, J.R. Pereira–Santos, S.P. Miranda, A. Rodrigues–Caetano, F.A. Gonçalves–Torres, 2007. Selection and testing of epiphytic yeasts to control anthacnose in post–harvest of papaya fruit. Scientia Horticulturae 111(2):179–185. [ Links ]
Deak, T. 1994. Rapid methods in food mycology. In: Spencer, R.C., Wright E.P., Newson S.W.B. (eds.), Rapid methods and automation in microbiology and inmunology. Intercept Press, Andover, pp 327–345. [ Links ]
Gariépy, T., C.A. Lévesque, S.N. de Jong, E. Rahe, 2003. Species specific identification of the Neofabraea pathogen complex associated with pome fruits using PCR and multiplex DNA amplification. Mycology Research 107(5): 528–536. [ Links ]
Guiraud, J.P. 1998. Microbiologi e alimentaire. 1st edition. Ed. Dunod, Paris. [ Links ]
Hirschhäuser, S., J. Fröhlich, 2007. Multiplex PCR for species discrimination of Sclerotiniaceae by novel laccase introns. International Journal of Food Microbiology 118(2): 151–157. [ Links ]
Hocking, A.D., S.L. Leong, B.A. Kazi, R.W. Emmett, E.S. Scott, 2007. Fungi and mycotoxins in vineyards and grape products. International Journal of Food Microbiology 119(1–2): 84–88. [ Links ]
Jellison, J., C. Jasalavich, 2000. A review of selected methods for the detection of degradative fungi. International Biodeterioration and Biodegradation 46(3): 241–244. [ Links ]
Johanson, A., H.C. Turner, G.J. McKay, A.E. Brown, 1998. A PCR–based method to distinguish fungi of the rice sheath–blight complex, Rhizoctoniasolani, R. oryzae and R. oryzae–sativae. FEMS Microbiology Letters 162(2): 289–294. [ Links ]
Kumpoun, W., Y. Motomura, Y. Harada, 2003. Inhibition of Aspergillus rot by sorbitol in apple fruit with water core symptoms. Postharvest Biology and Technology 29(2): 121–127. [ Links ]
Larone, D.H., 2002. Medically important fungi, a guide to identification. ASM Press. Washington D.C. [ Links ]
Lee, S.B., J.B. Taylor, 1990. Isolation of DNA from fungal mycelia and single spores. Academic Press, San Diego. [ Links ]
Marino, A., A. Nostro, C. Fiorentino, 2009. Ochratoxin A production by Aspergillus westerdijkiae in orange fruit and juice. International Journal of Food Microbiology 132(2–3):185–189. [ Links ]
NOM–111–SSA1–1994, Norma Oficial Mexicana, Bienes y Servicios. Método para la cuenta de mohos y levaduras en alimentos. In: http://www.salud.gob.mx/unidades/cdi/nom/111ssa14.html. [ Links ]
Ong, B.T., S.A.H. Nazimah, A. Osman, S.Y. Quek, Y.Y. Voon, D.M. Mat Hashim, P.M. Chew P.M., Y.W. Kong, 2006. Chemical and flavour changes in jackfruit (Artocarpus heterophyllus Lam.) cultivar J3 during ripening. Postharvest Biology and Technology 40(3):279–286. [ Links ]
Pitt, J.I., A.D. Hocking, 1999. Fungi and food spoilage. Springer Science Po., New York. [ Links ]
Roy, A.K., 1983. Perpetuation of Rhizopus artocarpi – the incitant of Rhizopus fruit rot of jack fruit (Artocarpus heterophyllus). Indian Phytopathology. 36(2):344–345. [ Links ]
SAGARPA, 2008. Anuario Estadístico de la Producción Agrícola de los Estados Unidos Mexicanos, México D.F. [ Links ]
Sambrook, J., D. W. Russell, 2001. Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor Laboratory Press, New York. [ Links ]
Saxena, A., B. A. Singh, P. Srinivas, 2008. Use of modified atmosphere packaging to extend shelf–life of minimally processed jackfruit (Artocarpus heterophyllus L.). Journal of Food Engineering 87(4): 455–466. [ Links ]
Shenasi, M., K.E. Aidoo, A.A.G. Candlish, 2002. Microflora of date fruits and production of aflatoxins at various stages of maturation. International Journal of Food Microbiology 79 (1–2):113–119. [ Links ]
Ton, H.T., M.K. Rok, K. Champada, 1990. Die–back: a new bacterial disease of champedakjack–fruit (Artocarpus sp. (Merr.) Thumb.). Journal of Thai Phytopathological Society 10(1–2):19–29. [ Links ]














