Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de micología
versión impresa ISSN 0187-3180
Rev. Mex. Mic vol.27 Xalapa dic. 2008
Contribuciones
Using ITS sequences to assess intraspecific genetic relationships among geographically separated collections of the myxomycete Didymium squamulosum
Uso de secuencias del ITS para determinar relaciones genéticas intraespecíficas entre colecciones separadas geográficamente del mixomicete Didymium squamulosum
Katherine E. Winsett1 * , Steven L. Stephenson1
1 Department of Biological Sciences, University of Arkansas Fayetteville, Arkansas 72701.
* Autor para correspondencia: kwinset@uark.edu
Recibido30 de marzo 2006
Aceptado 30 de agosto 2008
Resumen
Un análisis de las secuencias del ITS de Didymium squamulosum, un mixomicete reconocido como cosmopolita, muestra variación entre aislamientos geográficamente separados. Los ITS 1 y 2, y la región que codifica para el ADNr de la subunidad 5.8S fueron amplificados a partir de especímenes recolectados en localidades ampliamente dispersas. Las secuencias fueron analizadas a través de un análisis de parsimonia. Los árboles resultantes muestran clados separados, de moderadamente a bien soportados, que agrupan a algunas, pero no a todas las procedencias geográficas. El ITS es el primer marcador molecular que se examina para variación intra–específica en mixomicetes, y aunque el análisis de las secuencias muestra diferencias estadísticamente significativas entre las secuencias de los 14 especímenes considerados, la heterogeneidad en el ITS1 y en el ITS2 parece ser demasiado grande para esbozar cualquier conclusión biogeográfica significativa. La variación en la región 5.8S y la cantidad de variación encontrada en los ITS1 y 2 podría significar que los aislados examinados representan un complejo de especies gemelas, pero este marcador particular podría no ser útil para hacer esta distinción en este mixomicete.
Palabras clave: Region ITS, variación intraespecífica.
Abstract
An analysis of ITS sequences of Didymium squamulosum, a myxomycete regarded as cosmopolitan, shows variation among geographically separated isolates. ITS 1 and 2 and 5.8S rDNA were amplified from specimens collected in widely scattered localities. The sequences were analyzed by parsimony analysis and the resulting trees show separate, moderate and well supported clades grouping some, but not all, of the geographic locations. ITS is the first molecular marker examined for intraspecific variation in myxomycetes, and although sequence analysis shows statistically significant differences among sequences from the 14 specimens considered, the heterogeneity in ITS 1 and ITS 2 appears to be too great for any more meaningful biogeographical conclusions. Variation in the 5.8S and the amount of variation found in ITS 1 and 2 may signify that the isolates examined represent a complex of sibling species, although this particular marker cannot be used to make that distinction in this myxomycete.
Keywords: ITS, intraspecific variation
Introduction
The myxomycetes (also called plasmodial slime molds or myxogastrids) are a group of eukaryotic, phagotrophic bacteriovores usually present and often abundant in terrestrial ecosystems. The myxomycete life cycle involves two very different trophic stages, one consisting of uninucleate amoebae, with or without flagella, and the other consisting of a distinctive multinucleate structure, the plasmodium [8]. Under favorable conditions, the plasmodium gives rise to one or more fruiting bodies containing spores. The fruiting bodies of myxomycetes are somewhat suggestive of those produced by higher fungi, although they are considerably smaller (usually no more than 1–2 mm tall). The spores of myxomycetes are, for most species, apparently wind–dispersed and complete the life cycle by germinating to produce the uninucleate amoeboflagellate cells [14].
There are approximately 875 recognized species of myxomycetes [6]. The majority of these are probably cosmopolitan, but a few seem to be confined to the tropics or subtropics and some others have been collected only in temperate regions of the world [8,13,14]. Myxomycetes appear to be particularly abundant in temperate forests, but at least some species apparently occur in any terrestrial ecosystem with plants (and thus plant detritus) present [14,15].
Didymium squamulosum (Alb. & Shwein.) Fr. (Order Physarales, Family Didymiaceae) is an example of a truly cosmopolitan morphological species of myxomycete reported from numerous localities throughout the world (Figure 1) in habitats ranging from arctic tundra and subantarctic herbfields to lowland tropical rain forests and deserts [3,4,8,10]. The possible mechanism for long distance dispersal is the potential mobility of spores in this species and all known myxomycete species, which are small enough (8–11 µm in diameter [8]) to be transported by wind currents [14,17].
However, it is not yet known if this high dispersal potential, which in theory would allow for movement of spores between continents, is commonly realized. In fact, it has been hypothesized that this particular species of myxomycetes is not genetically ubiquitous. For example, results obtained from two studies [3,4] of the reproductive systems of Didymium squamulosum indicated that both nonheterothallic (presumably apomictic) and heterothallic strains of this species occurred in nature, suggesting that the Didymium squamulosum morphospecies is a complex of sibling biological species.
These data suggest there is much more information to be understood within the species that cannot be perceived by a consideration of morphology alone. The purpose of the study described herein was to obtain some initial baseline data on one example of a species (Didymium squamulosum) regarded as cosmopolitan, using DNA sequence analysis of a gene region with perceived utility for study of closely related myxomycetes [9]. It was anticipated that this type of analysis in the internal transcribed spacers of the ribosomal DNA (ITS rDNA) region of Didymium squamulosum would provide evidence of any underlying genetic diversity and possible biogeographic patterns that may exist in cosmopolitan myxomycetes, thus representing a source of data by which to study mechanisms of dispersal, population biology and evolution within myxomycetes.
Materials and methods
DNA amplification and sequencing
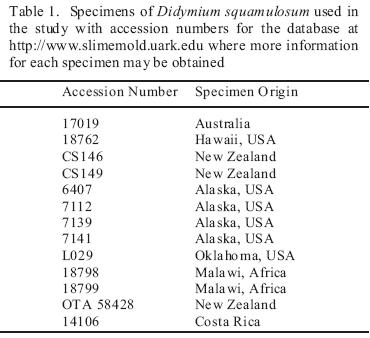
Fourteen specimens (fruiting bodies from moist chamber culture or field collections) of Didymium squamulosum from widely scattered localities throughout the world (Table 1) were analyzed for biogeographically meaningful variation in the internal transcribed spacers 1 and 2 (ITS1, ITS2). For DNA extraction, 4–6 sporangia were ground in Sigma acid–washed glass beads with a sieve size of 710–1180 m to release the spores. DNA was extracted from the spores using 150 L of 5% Chelex suspension at 56 C for 4 hours then 98 C for 30 minutes in a Bio–Rad PTC–0200 DNA Engine Thermal Cycler. ITS 1 and 2, 5.8S, and flanking small and large subunit (SSU and LSU) regions of rDNA were amplified by means of the polymerase chain reaction (PCR) using amplification primers in the SSU and LSU as described by Martín et al. [9](Table 2). The DNA templates were amplified in Invitrogen Platinum Blue PCR SuperMix with the primers using a PCR protocol, which consisted of an initial activation reaction at 94 °C for 2 minutes and 35 repetitions of 94 °C for 30 sec, 52 °C for 1 min, and 72 °C for 2 min. The PCR products were transformed and cloned, using the methods of the Invitrogen TOPO TA Cloning Kit for Sequencing. Four clones were chosen for sequencing in an effort to assess any intra–clonal variation. DNA for sequencing was extracted with the Qiagen QIAprep Spin Miniprep Kit and sequenced on a Perkin–Elmer ABI 3700 sequencer. Internal sequencing primers in the 5.8S were designed specifically for Didymium squamulosum (Table 2).
Sequence alignment and analysis
The sequences were initially aligned using ClustalX [18] and manually edited in SeaView [5]. Thirteen isolates were analyzed by Maximum Likelihood analysis of the homologous alignment of sequences in PAUP 4.0 [16] using the likelihood settings from the best–fit model (TVMef+I+G)
Results
The ITS region from 13 isolates of Didymium squamulosum was amplified using known primers from Martín et al [9], which are in flanking regions of the ITS 1 and 2 in the small and large subunits, thus adding approximately 132 characters to the sequence. Internal primers in the 5.8S were developed for sequencing purposes using known sequences of Didymium squamulosum. The sequenced region for every isolate includes all of ITS 1, 5.8 S and ITS2 rDNA, and length variation of the sequences among the specimens was evident in both ITS 1 and ITS2, with the total amplified region ranging from approximately 1090 to 1416 base pairs. In each sequence, approximately 302 base pairs come from the rDNA genes (26S, 5.8S and 19S), thus leaving a sequence length of 788–1114 base pairs as the total length of ITS 1 and 2. The sequence alignment offered 397 unambiguously aligned sites (22.8% of the total length), and of these, only 95 sites (5.45% of the total length) are within either ITS 1 or ITS2. A graphical representation of the sequence alignment using Fingerprint [7] illustrates the heterogeneity of the full sequences amplified for this species (Figure 2).
The maximum likelihood phylogram displays the relationships between isolates based upon analysis using the best–fit model selected by AIC in ModelTest. Bootstrap values greater than 60 are displayed (Figure 3). Well supported groups of isolates are found in the data analysis. Two of the well supported groups are made up of isolates found at the same collecting sites: (a) New Zealand 26852, 26833 and (b) Alaska 7112, 7139, 7141. They are supported with bootstrap values of 92 and 94 respectively. However, a fourth isolate from Alaska and a third isolate from New Zealand do not group with the other isolates from the geographic region.
Discussion
Didymium squamulosum as a study organism
Didymium squamulosum is known to be a widely distributed species with variable morphological features [3,4]. In this species, as with many species of myxomycetes, it is not known if morphological variation is an expression of phenotypic plasticity or phenotypic evidence of genetic variation or divergence [2]. A study of sporangium characteristics for isolates of Didymium squamulosum did not coincide with genetic variation measured by isozyme variation and the results of plasmodium fusion tests [3,4]. These previous studies suggest a complex of biological species within the recognized morphological species of Didymium squamulosum. Thus, the genetic variation within Didymium squamulosum is potentially a very valuable tool to understand the intraspecific diversity and the potential cryptic species–level diversity within the morphological species.
ITS as a genetic marker for geographic patterns
Internal transcribed spacers (ITS) are intervening sequences between the small and large subunits of the ribosomal DNA (rDNA). In Physarum polycephalum, rDNA occurs on a single, linear, extrachromosomal DNA molecule in the nucleolus of the cell. The large and small subunit exons are separated by two spacers and the 5.8S exon. There are two copies of each region arranged as a palindrome around a large central spacer [1, 12].These two spacers, ITS 1 and ITS 2 were successfully amplified for an intrageneric study of Lamproderma [9]. Interestingly, it was reported that the alignment for this region was unambiguous for the 3 species investigated [9]. This sequence, though, was problematic in Didymium squamulosum due to the extensive variation in both nucleotide and length diversity within the species limiting the usefulness of the marker for genetic study within populations. In this baseline data set, less than 6% of the ITS sequence was found to be homologous.
The analysis of the ITS1–5.8S–ITS2 regions carried out in the present study divided the collections of Didymium squamulosum into clades with some evidence of a geographical pattern. Collections from the same region (e.g., two specimens from New Zealand and three from Alaska) share a clade in the topology of the tree. However, the ITS complex is too variable to give a reliable description of the relationships among geographically disjunct populations. The majority of unambiguously aligned sites for analysis were chosen not from ITS1 or ITS2, but from the flanking large and small subunit rDNA and the 5.8S segments. The ITS regions themselves are too variable in nucleotide content and length to construct an unambiguous alignment.
Biogeographical inference
The topology of the tree from these analyses does separate the samples into well–supported clades. However, the placement of isolates in the tree does not, in general, follow a discernable biogeographical pattern (see for example, Alaska 6407, Costa Rica 14106), which may also be a result.
The data obtained thus far indicate that additional study of the relationships among specimens of Didymium squamulosum and other myxomycetes is warranted. The variation uncovered in the ITS sequences of Didymium squamulosum suggest that a more complete genetic profile within the species will offer data to explore local and global dispersal patterns beyond the variation observed in the morphological characteristics of the species. However, ITS seems to have limited usefulness for investigations within a species due to extensive heterogeneity of sequences between isolates (Figure 2). It will be important to isolate a gene or gene region that will offer enough variation within a species without presenting too much and unsuitable variation for intraspecific analysis.
Acknowledgements
This research project was funded in part by two grants (DEB–0316284 and DEB–0329102) from the National Science Foundation. Thanks are also due to Dr. Satyendra Rajguru and Dr. Jeffrey Silberman for technical and scientific assistance and to reviewers for their helpful comments.
1. Campbell, G.R., V.C. Littau, P.W. Melera, V.G. Allfrey, E.M. Johnson. 1979. Unique sequence arrangement of ribosomal genes in the palindromic rDNA molecule of Physarum polycephalum. Nucleic Acids Research 6:1433–1447. [ Links ]
2. Clark, J., 2000. The species problem in the myxomycetes. Stapfia 73: 39–53. [ Links ]
3. Clark, J., S.L. Stephenson, 2003. Biosystematics of the myxomycetes Didymium squamulosum, Physarum compressum, and Physarum melleum: additional isolates. Mycotaxon 85:85–89. [ Links ]
4. El Hage, N. C. Little, J.D. Clark, S.L. Stephenson, 2000. Biosystematics of the Didymium squamulosum complex. Mycologia 92:54–64. [ Links ]
5. Galtier, N., Gouy, M., Gautier, C., 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Computer Applications in the Biosciences 12: 543–548. [ Links ]
6. Lado, C., 2001. Nomenmyx. A Nomenclatural Taxabase of Myxomycetes. Cuad. Trab. Fl. Micología Ibérica 16:1–221. [ Links ]
7. Lou, M., G.B. Golding. 2007. Fingerprint: visual depiction of variation in multiple sequence alignments. Molecular Ecology Notes 7: 908–914. [ Links ]
8. Martin, G. W., Alexopoulos, C. J., 1969. The Myxomycetes. University of Iowa Press, Iowa City. [ Links ]
9. Martín, M.P., C. Lado, S. Johansen., 2003. Primers are designed for amplification and direct sequencing of ITS region of rDNA from Myxomycetes. Mycologia 95: 474–479. [ Links ]
10. Pickering, J. 2006. Global Mapper. News report for International Biogeography Society. March 2006. (http://www.discoverlife.org/pa/or/polistes/fe/2006ibs.html). [ Links ]
11. Posada, D., K.A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818. [ Links ]
12. Seebeck, T., J. Stalder, R. Braun. 1979. Isolation of a minichromosome containing the ribosomal genes from Physarum polycephalum. Biochemistry 18:484–490. [ Links ]
13. Stephenson, S. L., Schnittler, M., Lado, C., Estrada–Torres, A., Wrigley de Basanta, D., Landolt, J. C., Novozhilov, Y. K., Clark, J., Moore, D. L., Spiegel, F. W., 2004. Studies of neotropical mycetozoans. Systematics and Geography of Plants 74: 87–108. [ Links ]
14. Stephenson, S. L., Schnittler, M., Novozhilov, Y.K., 2008. Myxomycete diversity and distribution from the fossil record to the present. Biodiversity and Conservation 17:285–301. [ Links ]
15. Stephenson, S. L., Stempen, H., 1994. Myxomycetes: a Handbook of Slime Molds. Timber Press. [ Links ]
16. Swofford, D. L., 2002. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts. [ Links ]
17. Tesmer, J., Schnittler, M. 2007. Sedimentation velocity of myxomycete spores. Mycological Progress 6:229–234. [ Links ]
18. Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G., 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: [ Links ]














