Introduction
Molecular fouling is a ubiquitous phenomenon in aquatic ecosystems and aquaculture systems. When surfaces are submerged in seawater, dissolved organic matter adheres to these surfaces in just seconds, in a process termed molecular biofouling, forming a thin film (≈100 nm) that promotes the colonization of various microorganisms from diverse taxa (Qian et al. 2007). Consequently, molecular fouling is rapidly followed by a biofouling process, in which these microorganisms begin to form a microbial community or biofilm. A microbial biofilm could be defined as an aggregate of diverse microorganisms belonging to diverse domains embedded within a self-produced matrix of extracellular polymeric substance adhered to each other and/or to a surface (Vert et al. 2012).
Both phototrophic- and heterotrophic-biofilm-based systems have been successfully used in aquaculture (Ballester et al. 2007, Becerra-Dorame et al. 2011) for diverse purposes, including bioremediation, probiotic activity, and, more recently, as a direct food source (Martínez-Córdova et al. 2014). Recent reports revealed that the use of these microbial communities may improve not only the water quality in shrimp culture systems, but also the growth performance and the physiological conditions of shrimp (Martínez-Córdova et al. 2014, Sruthisree et al. 2015). There is evidence that biofilms are constituted by bacteria, archaea, microalgae, organic and inorganic matter, and exopolymeric substances, as well as other microorganisms in lower proportions (Tolker-Nielsen and Molin 2000). From these, bacteria are perhaps the most important constituents because of their roles in the adherence of the biofilm to the surface, and in the complex interaction network occurring within the microbial community (Avendaño-Herrera and Riquelme 2007, Roeselers et al. 2008).
Several studies have been performed to elucidate the bacterial diversity of biofilms (Tolker-Nielsen and Molin 2000, Donlan 2002). Most bacteria thriving in biofilm microbial communities are usually identified through microscopic, biochemical, or molecular-based methods, such as restriction fragment length polymorphism (RFLP), denaturing gradient gel electrophoresis (DGGE), and other approaches (Bourne et al. 2004, Becerra-Dorame et al. 2011). Culture-dependent methods have been useful for the study of bacterial communities; however, the diversity of microorganisms in aquaculture has been elucidated in a very small fraction of bacterial communities, because 85-99% of the bacteria from the marine environment are not readily culturable or are simply unculturable (Martinez-Porchas and Vargas-Albores 2015). On the other hand, RFLP and DGGE reveal the presence of a major proportion of some species, but with lower resolution in terms of detectable limits compared to next-generation sequencing (NGS) technologies (Al-Awadhi et al. 2013).
A higher resolution of the bacterial community composition in biofilms used for aquaculture purposes could provide useful information, considering that bacteria-bacteria and (micro)algae-bacteria interactions include signal transduction, gene transfer, and nutrient exchange (Kouzuma and Watanabe 2015), which contributes to the success of these microbial communities. Moreover, this information may reveal differences in the taxonomic profile between both autotrophic- and heterotrophic-based biofilms, which is also relevant from a bioremediation perspective since microbial conglomerates take up and recycle nutrients or “wastes” produced by cultured organisms.
NGS is a useful tool for obtaining this information and could provide an insight into the bacterial diversity of the microbial consortia through analysis of the hypervariable regions of the prokaryote 16S rRNA genes (Martinez-Porchas and Vargas-Albores 2015). 16S rRNA genes have at least one copy per genome, and 9 hypervariable regions have been detected that show considerable sequence diversity among bacteria. For instance, NGS has been used to study bacterial communities (including biofilms) in urban drinking water (Wu et al. 2015) and biofilms detected in fish processing plants (Langsrud et al. 2016), but not yet in marine biofilms with aquaculture purposes.
The aim of this study was to evaluate the taxonomic profiles of the bacterial communities detected, with 16S rRNA sequences, in mature phototrophic and heterotrophic marine biofilms used for aquaculture, to elucidate the bacterial community structure of these conglomerates.
Materials and methods
Biofilm culture
Autotrophic and heterotrophic biofilms were promoted and matured into six 200-L plastic tanks. These types of biofilms have been previously used as a direct food source for shrimp cultured under laboratory conditions with successful results (Becerra-Dorame et al. 2011, Becerra-Dorame et al. 2014).
The conditions for the proliferation of the biofilms were as follows: tanks were filled with 180 L of marine water taken from an estuary and previously filtered through a sand filter. Constant aeration was provided by diffusers placed at the bottom of the tanks. Artificial substrates consisting of plastic mesh-16 were placed vertically into each tank, and a surface:volume ratio of 5.6 m2 of substrate per square meter of water was achieved. Thereafter, tanks were randomly divided into groups to test 2 different types of biofilms (3 tanks/biofilm type): phototrophic biofilm (PAb) and a heterotrophic biofilm (Hb). The formation of both biofilms was based on unspecific marine microorganisms already present in the marine water.
For PAb, a carbon:nitrogen ratio of 5:1 was maintained in the water by the periodic addition of Triple-17 fertilizer (17% nitrogen, 17% phosphorous, 17% potassium). The quantification of carbon and nitrogen was performed spectrophotometrically (Hanna Instruments, NE, USA) to calculate the amount of fertilizer to be added. The tanks were exposed to outdoor conditions, including sunlight, and inoculated with 1 L (1 × 106 cells/mL) of the marine benthic diatom Navicula sp. Temperature was maintained at 30 ºC, dissolved oxygen at 4-7 mg/L, pH at 8.5-9.0, and light intensity at 340-1,020 umol/m2·s. The diatom Navicula sp. was previously grown under laboratory conditions using 1 L Erlenmeyer flasks with sterile marine water and f/2 media (Guillard 1975) and was used as the biofilm promoter (Martínez-Córdova et al. 2017).
For Hb, a carbon:nitrogen ratio of 20:1 (Avnimelech 1999) was used. Briefly, molasses was added as a carbon source to maintain an organic carbon concentration of ~20 mg/L. The organic carbon levels were monitored by a Torch Combustion TOC Analyzer (Teledyne-Tekmark; OH, USA). Temperature and dissolved oxygen were set to similar levels as those for the PAb, but the pH range was 7.5-8.0. A carbon:nitrogen:phosphorous ratio of 20:3:1 was maintained by adding 2 mL molasses, 2 g NaNO3 and (NH4)2SO4, and 0.05 g K2HPO4.
Biofilms were allowed to grow for 30 days prior to the trial. Water loss due to evaporation prior and during the trial was compensated for using sterile and ultrapure freshwater previously autoclaved and treated with a Milli-Q integral water purification system (Millipore; MA, USA) to avoid contamination of the system with other live microbes. Once the biofouling occurred and biofilms were attached to the substrates (~40-60 mg of biofilm per square centimeter), samples were taken at 0, 15, and 30 days.
Taxonomic profile of bacterial communities
Bacteria present in the biofilms were detected through a targeted-metagenomic approach using the high throughput sequencing of the 16S rRNA gene, which was used as a taxonomic biomarker.
Samples were collected from each treatment and the DNA was isolated. Briefly, samples were obtained by scratching the surface of the plastic substrate and washed with sterile, nuclease-free water (Promega, USA) to eliminate possible unattached-planktonic bacteria. Samples from each biofilm group were pooled and homogenized at 6 m/s for 30 s using a FastPrep-24 5G homogenizer (MP Biomedicals; CA, USA).
Randomly-sheared, high-molecular-weight metagenomic DNA (free of PCR inhibitors) directly from culturable and unculturable bacteria was isolated following the specifications of a commercial kit (Power Biofilm DNA isolation kit, MO BIO Laboratories; CA, USA). The DNA quality was monitored by a microfluidic electrophoresis instrument (2200 Tapestation, Agilent; CA, USA). Briefly, 1 µL of the DNA isolate was mixed with 10 µL of the gDNA sample buffer (Agilent). Of this mixture, 1 µL per sample was added to a microfluidic chip (gDNA ScreenTape, Agilent) to assess the quantity and quality of DNA (range: 200 to >60,000 bp DNA fragments). The gDNA Ladder (Agilent) was used as the reference. Samples with DNA integrity numbers above 7 were considered for the construction of a library. Additionally, DNA was extracted following the specifications of the Meta-G-Nome DNA isolation kit (Epicentre; CA, USA) from the marine water used to grow both biofilms.
Library preparation
16S-metagenomic library preparation was carried out following the “16S-metagenomic sequencing library preparation guide” (Illumina 2013). In short, the protocol targets the 16S V3 and V4 regions together using the primers reported by Klindworth et al. (2012), modified with sequencing adapters and dual index barcodes: 16S Overhang adapter/Forward Primer: 5' TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG 3'; 16S Overhang adapter/Reverse Primer: 5' GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3'.
The first amplification of the V3/V4 region was carried out in 25 µL reactions using 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems; MA, USA) under the following thermal cycling conditions: initial denaturation at 95 ºC for 3 min; 25 cycles of 95 ºC for 10 s, 55 ºC for 30 s, 72 ºC for 30 s; and a final extension at 68 ºC for 5 min. Amplicons ranging from 450 to 550 bp were purified using magnetic AMPure XP beads (Beckman Coulter; FL, USA) to separate the 16S V3/V4 amplicon from free primers and primer dimer species, following the manufacturer’s specifications. After isolation, the amplicons were attached with dual indexes and sequencing adapters using the Nextera XT Index Kit (Illumina; CA, USA) and 2× KAPA HiFi HotStart ReadyMix. The above thermal cycling conditions were performed for 8 cycles, and the resulting PCR products were purified as mentioned above.
Finally, the resulting library was quantified and the quality checked by electrophoresis (2200 Tapestation, Agilent; CA, USA) using microfluidic chips (with an analysis range of 35 to 1,000 bp fragments, D1000 ScreenTape, Agilent; CA, USA). Briefly, 2 µL of the indexed-library was mixed with 2 µL of High Sensitivity D1000 buffer. The samples were finally loaded into a High Sensitivity D1000 ScreenTape and analyzed by electrophoresis (2200 Tapestation).
Sequencing
Libraries were diluted to 4 nM using 10 mM Tris (pH 8.5) and pooled. Thereafter, libraries were denatured with 0.2 N NaOH. A standard PhiX Control Library (Illumina), which is a well-defined short genome sequence that allows for the calculation of error rates, was also denatured and used as an internal control.
Thereafter, both denatured libraries (sample and control) were adjusted to a final concentration of 8 pM and mixed (95% library + 5% PhiX Control). Finally, the mixture was heated to 96 ºC for 2 min, immediately cooled in ice for 5 min, and loaded into a MiSeq v3 Reagent Tray (Illumina). The tray was inserted into a MiSeq sequencing instrument (Illumina), which contained a MiSeq v3 Flow Cell (Illumina) with a capacity of 25 million reads. Results were obtained after 602 (2 × 301) cycles. Artificial replicate sequences were removed and the resulting paired-end reads were assembled. Thereafter, sequences were submitted to a dereplication process, followed by length (2.0 standard deviations) and ambiguous base filtering (max ambiguous bp = 5).
Data analysis
Sequences were submitted to the MG-RAST server platform and the default options for low-quality reads trimming and sequence screening was performed. Sequences outside the range of ±2 standard deviations were not considered for the analysis. The taxonomic classification and diversity analyses were performed by comparison against the SILVA (128) database using the MG-RAST pipeline, where tasks for detection, clustering at 97% similarity and RNA identification, annotation mapping, and abundance profiles were used. Principal component analysis was performed using the STAMP (statistical analysis of taxonomic and functional profiles) software (Parks et al. 2014). A repeated-measures analysis of variance was used for data analysis, considering relative abundance and time as factors, and a significance value of P = 0.05. Finally, alpha and beta diversities were calculated using an analytic hierarchy method as described by Goepel (Goepel 2013).
Results
DNA extracted and purified from PAb and Hb registered DNA integrity number values above 8.4 and 7.4, respectively. After amplification, amplicons with sizes 501 to 540 bp were obtained for both biofilms during the trial; the percentages of integrated area of these amplicons were ≥90.1 and ≥89.4%, for PAb and Hb, respectively, whereas peak molarity values ranged from 3,050 to 3,700 pmol/L
Averages of 1,010,601 and 496,259 reads were classified up to a taxonomic level in the PAb and Hb samples, respectively. Of these, ~35-40% were classified up to genus or species level. Bacterial diversity remained constant in both biofilms with respect to time (with a few exceptions); on the other hand, the relative abundance of the most representative bacteria did not register significant changes, except for a few specimens that varied over time (P < 0.05) (Table 1, Fig. 1). Herein, alpha diversity changed over time, revealing a loss of species in the PAb, and a gain of species in the case of the Hb (Table 2). Proteobacteria represented the most abundant phylum in both biofilms during the experimental period. Herein, 48% and 36% of the analyzed reads of PAb and Hb during the 3 sampling dates corresponded to Proteobacteria (Fig. 1). Most of these reads were assigned to the Alpha-, Beta-, or Gamma-Proteobacteria class in the 2 different biofilms. After Proteobacteria, Chlamidiae-Verrucomicrobia (11%), Bacteriodetes (8%), and Planctomycetes (5%) were the most abundant phyla in the PA biofilm, whereas Planctomycetes (22%), Bacteriodetes (8%), Actinobacteria (6%), and Chamydiae (2%) were found in the Hb (Fig. 1). Several other phyla were detected in both biofilms but in lower proportions (<1%).
Table 1 Proportion of bacterial species detected in the phototrophic (PAb) and heterotrophic (Hb) biofilms over time (0, 15, and 30 d). Proportions are based on the total reads assigned up to species level. P values for biofilm type and time factors are also shown.
| Phylum | Species detected in marine water | PAb 0 |
PAb 15 |
PAb 30 |
Hb 0 |
Hb 15 |
Hb 30 |
P value (biofilm/time) |
| Proteobacteria | Ketogulonicigenium vulgare | 37.9 | 38.9 | 43.1 | 27.8 | 25.5 | 20.5 | (0.00/0.15) |
| Rhodobacter capsulatus SB 1003 | 8.2 | 9.5 | 6.2 | 0.0 | 0.0 | 0.0 | (0.00/0.00) | |
| Alteromonas macleodii | 1.0 | 0.0 | 0.8 | 0.0 | 0.0 | 0.0 | (0.00/0.00) | |
| Verminephrobacter eiseniae | 0.5 | 1.0 | 0.7 | 9.5 | 12.3 | 8.3 | (0.00/0.09) | |
| Sulfiromonas autotrophica | 3.3 | 1.5 | 1.1 | 5.5 | 2.7 | 2.5 | (0.02/0.06) | |
| Nitrosospira multiformis | 3.0 | 2.5 | 2.0 | 0.0 | 0.0 | 0.0 | (0.00/0.22) | |
| Micavibrio aeruginosavorus | 0.0 | 0.0 | 0.0 | 4.2 | 5.1 | 2.4 | (0.00/0.11) | |
| Arcobacter sp. L | 0.0 | 0.0 | 0.0 | 1.0 | 3.5 | 1.3 | (0.00/0.04) | |
| Hyphomonas neptunium | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 | 0.8 | (0.00/0.01) | |
| Alcanivorax dieselolei | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Desulfomonile tiedjei | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| alpha proteobacterium HIMB59 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Ruegeria pomeroyi | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Parvularcula bermudensis | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Vibrio sp. | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Chlamydiae | Simkania negevensis Z | 10.1 | 9.2 | 15.2 | 1.0 | 1.0 | 1.2 | (0.00/0.18) |
| Parachlamydia acanthamoebae UV-7 | 0.5 | 0.5 | 1.7 | 1.0 | 0.5 | 2.2 | (0.03/0.04) | |
| Waddlia chondrophila | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | - | |
| Bacteriodetes | Haliscomenobacter hydrossis DSM 1100 | 12.3 | 15.5 | 9.1 | 27.7 | 29.1 | 20.5 | (0.02/0.30) |
| Echinicola vietnamensis DSM 17526 | 7.5 | 8.0 | 4.5 | 0.0 | 0.0 | 0.0 | (0.00/0.05) | |
| Riemerella anatipestifer | 0.0 | 0.0 | 0.0 | 2.2 | 0.5 | 0.9 | (0.00/0.03) | |
| Robiginitalea biformata | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Cytophaga hutchinsonii | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Verrucomicrobia | Opitutus terrae PB90-1 | 2.0 | 1.5 | 4.1 | 0.5 | 0.5 | 1.5 | (0.01/0.04) |
| Planctomycetes | Planctomyces brasiliensis DSM 5305 | 6.0 | 3.5 | 4.1 | 2.1 | 4.0 | 1.4 | (0.01/0.01) |
| Rhodopirellula baltica SH 1 | 5.5 | 4.5 | 3.8 | 0.0 | 0.0 | 0.0 | (0.00/0.20) | |
| Phycisphaera mikurensis | 0.0 | 0.0 | 0.0 | 1.0 | 0.5 | 1.6 | (0.00/0.02) | |
| Firmicutes | candidate division WWE3 bacterium | 0.0 | 1.0 | 1.2 | 0.0 | 0.0 | 0.0 | (0.00/0.00) |
| Streptococcus agalactiae | 0.0 | 0.5 | 0.4 | 0.0 | 0.5 | 1.2 | (0.00/0.00) | |
| Saprospira grandis | 1.0 | 1.0 | 0.5 | 0.0 | 0.0 | 0.0 | (0.00/0.54) | |
| Acetohalobium arabaticum | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 1.4 | (0.00/0.00) | |
| Halobacterioides halobius | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | - | |
| Elusimicrobia | Elusimicrobium minutum | 0.0 | 0.1 | 0.3 | 0.5 | 0.2 | 1.1 | (0.01/0.03) |
| Actinobacteria | Microbacterium testaceum | 0.0 | 0.0 | 0.0 | 13.1 | 11.3 | 23.1 | (0.00/0.05) |
| Tropheryma whipplei | 0.0 | 0.0 | 0.0 | 1.2 | 1.0 | 5.7 | (0.00/0.02) | |
| Propionibacterium acidipropionici | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.5 | (0.00/0.00) | |
| Other | Other | 1.2 | 1.3 | 1.0 | 0.4 | 1.8 | 1.9 | (0.05/0.06) |
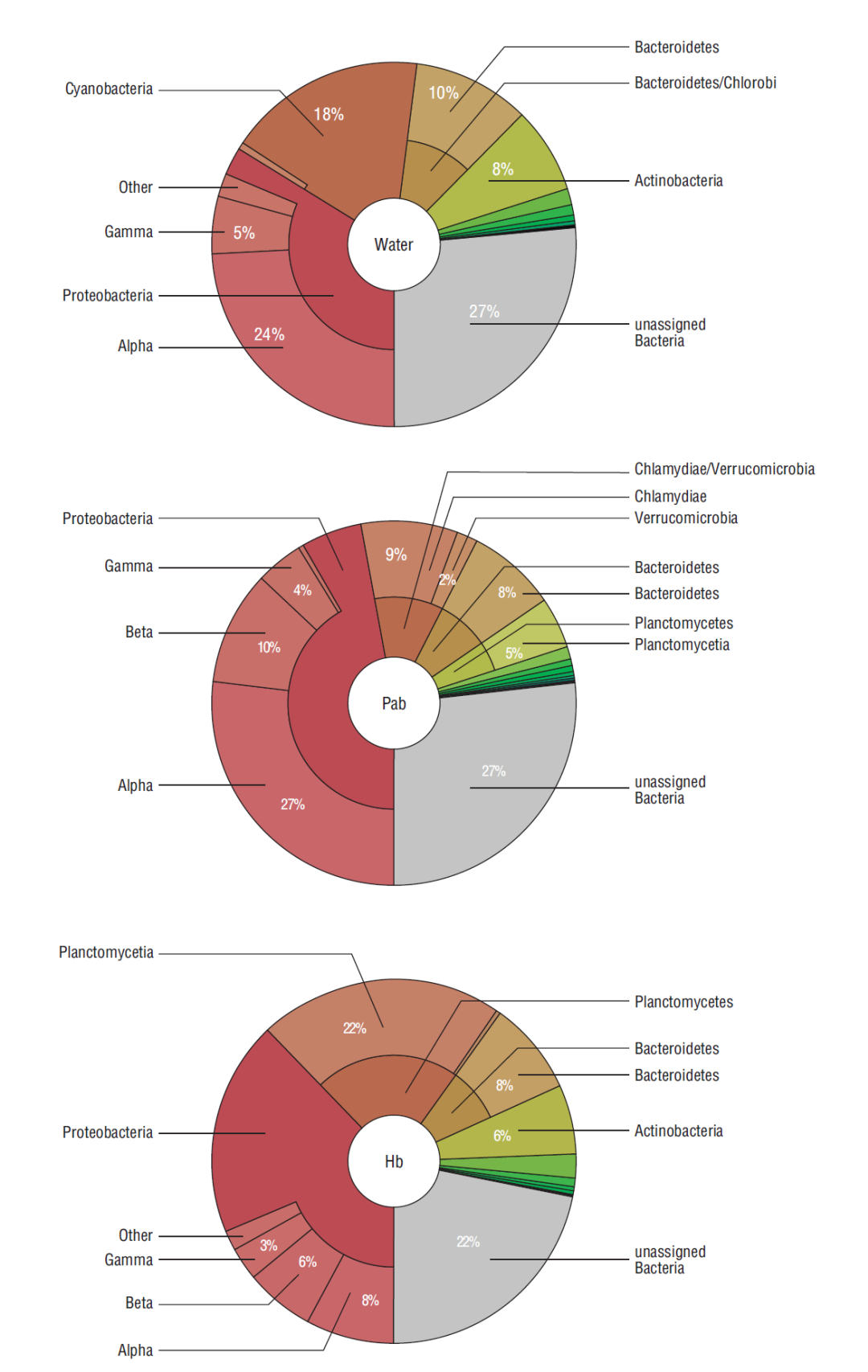
Figure 1 Taxonomic classification (phylum and class) of bacteria detected in water used for the biofilm culture, in the phototrophic biofilm (PAb) and in the heterotrophic biofilm (Hb).
Table 2 Comparison matrix containing the alpha (α) and beta (β) diversity values of phototrophic (PAb) and heterotrophic (Hb) biofilms at 0, 15, and 30 days of culture.
| PAb 0 | PAb 15 | PAb 30 | Hb 0 | Hb 15 | Hb 30 | |
| β-Diversity | ||||||
| PAb 0 | 0.00 | 0.01 | 0.02 | 0.26 | 0.25 | 0.29 |
| PAb 15 | 0.01 | 0.00 | 0.02 | 0.25 | 0.24 | 0.28 |
| PAb 30 | 0.02 | 0.02 | 0.00 | 0.28 | 0.27 | 0.29 |
| Hb 0 | 0.26 | 0.25 | 0.28 | 0.00 | 0.02 | 0.05 |
| Hb 15 | 0.25 | 0.24 | 0.27 | 0.02 | 0.00 | 0.05 |
| Hb 30 | 0.29 | 0.28 | 0.29 | 0.05 | 0.05 | 0.00 |
| α-Diversity | 1.98 | 1.90 | 1.85 | 1.86 | 1.91 | 2.05 |
Different bacterial profiles, in terms of relative abundance and detection of particular species, were observed in the 2 different biofilms (Fig. 2). Of the reads assigned to the species level, Ketogulonicigium vulgare, Rhodobacter capsulatus, Nitrosospira multiformis, Sulfiromonas autotrophica (all Proteobacteria), Simkania negevensis (Chlamydiae), Haliscomenobacter hydrossis, Echinicola vietnamensis (Bacteriodetes), Opitutus terrae (Verrucomicrobia), Planctomyces brasiliensis, and Rhodopirellula baltica (Planctomycetes) accounted for 89-94% of the reads in PAb during the trial (Table 1). For the Hb, of the reads assigned to the species level, K. vulgare, Verminephrobacter eiseniae, S. autotrophica (all Proteobacteria), H. hydrossis (Bacteriodetes), P. brasiliensis (Planctomycetes), Microbacterium testaceum, and Tropheryma whipplei (Actinobacteria) accounted for 83-93% of the reads (Table 1).
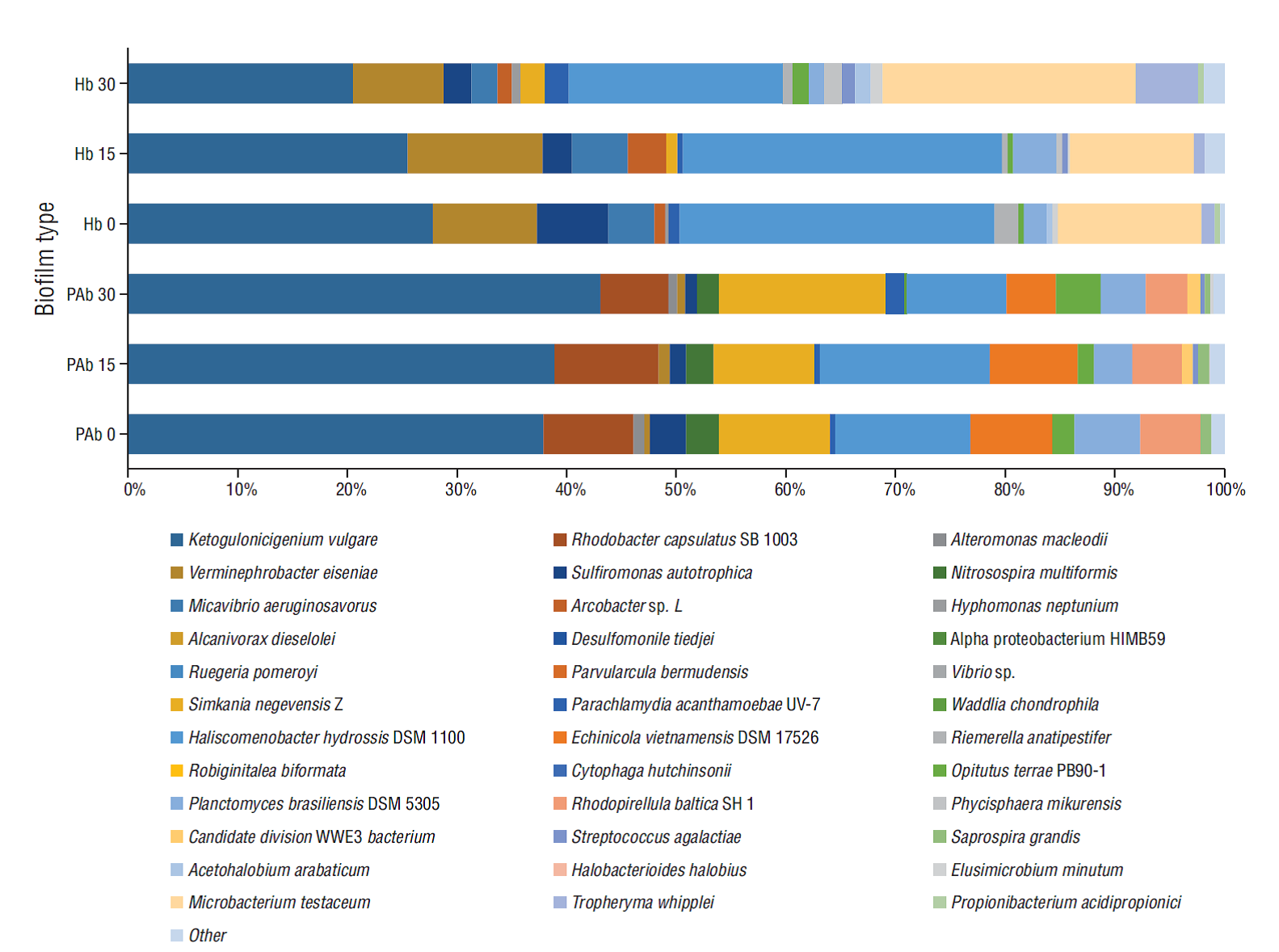
Figure 2 Bacterial profiles of phototrophic (PAb) and heterotrophic (Hb) biofilms over time (0, 15 and 30 days). Proportions are based on the total number of reads assigned up to species level.
All the species detected in both biofilms were first detected in water in different proportions. Of the 36 species detected in water (with at least 1% of the reads classified to species), 27 were detected in both biofilms from at least one sampling date (Table 1). However, beta diversity analysis together with PCA revealed that the weighted taxonomic profile of both biofilms was diametrically different (Table 2, Fig. 3).
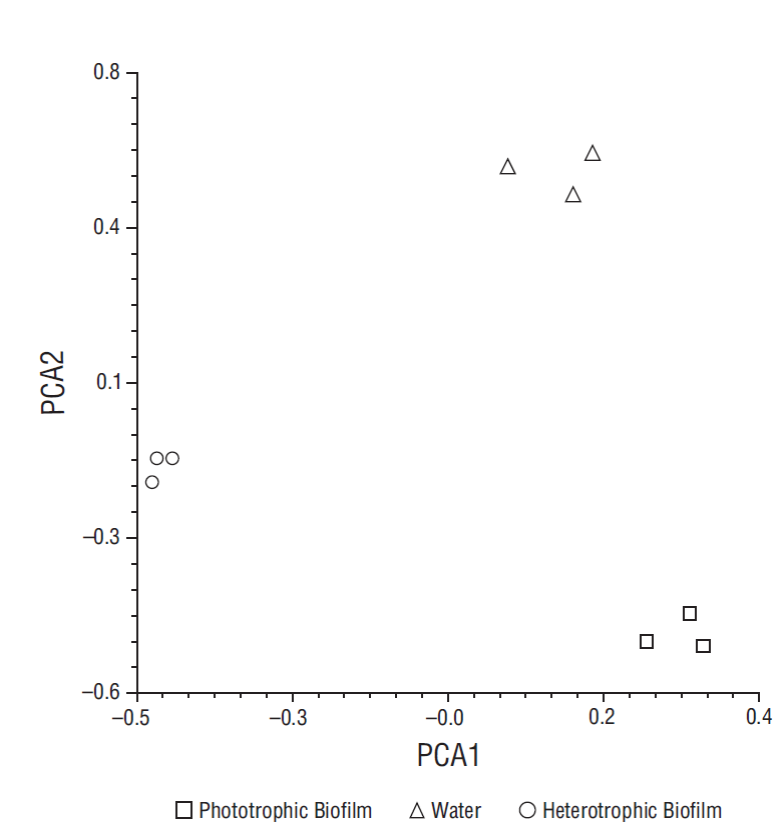
Figure 3 Principal component plot illustrating differences in bacterial community structure of heterotrophic and phototrophic biofilms after 30 days, considering time (PCA1 < 10%) and sample type (PCA2 > 85%).
Some species were detected in all biofilm samples and others were exclusive for one biofilm type. For instance, K. vulgare, V. eiseniae, S. negevensis, H. hydrossis, P. brasiliensis, and Streptococcus agalactiae were detected in both phototrophic and heterotrophic biofilms (Table 3), with K. vulgare (21-43%) and H. hydrossis (9-29%) detected in high proportions in both biofilms.
Table 3 Species detected in both biofilms, in only the phototrophic biofilm (PAb only), and in only the heterotrophic biofilm (Hb only).
| Phylum | Detected in both biofilms | PAb only | Hb only |
| Proteobacteria | Ketogulonicigenium vulgare | ||
| Verminephrobacter eiseniae | |||
| Sulfiromonas autotrophica | |||
| Nitrosospira multiformis | |||
| Micavibrio aeruginosavorus | |||
| Arcobacter sp. L | |||
| Hyphomonas neptunium | |||
| Chlamydiae | Simkania negevensis | ||
| Parachlamydia acanthamoebae | |||
| Bacteriodetes | Parachlamydia hydrossis | ||
| Echinicola vietnamensis | |||
| Riemerella anatipestifer | |||
| Planctomycetes | Planctomyces brasiliensis | ||
| Rhodopirellula baltica | |||
| Phycisphaera mikurensis | |||
| Firmicutes | Candidate division WWE3 bacterium | ||
| Streptococcus agalactiae | |||
| Saprospira grandis | |||
| Acetohalobium arabaticum | |||
| Actinobacteria | Microbacterium testaceum | ||
| Propionibacterium acidipropionici |
Regarding bacteria exclusively detected in the phototrophic biofilm, species such as N. multiformis, E. vietnamensis, R. baltica, Saprospira grandis, and a candidate division WWE3 bacterium were observed (Table 3), whereas Micavibrio aeruginosavorus, Arcobacter sp. L, Hyphomonas neptunium, Riemerella anatipestifer, Phycisphaera mikurensis, Acetohalobium arabaticum, T. whipplei, M. testaceum, and Propionibacterium acidipropionici were exclusively detected in heterotrophic biofilms (Table 3).
Discussion
The results revealed that bacterial composition in terms of diversity and relative abundance was consistent over time for both biofilms. It is likely that the proliferation period prior to the trial (30 days) was sufficient for the microbial conglomerates to achieve a mature phase where the biofilm composition was less variable over time. Moreover, the constant experimental conditions could have contributed to maintaining a uniform composition.
Bacteria belonging to the Proteobacteria, Bacteroidetes, and Planctomycetes phyla seemed to form the core structure of both Hb and PAb (also Chlamydiae). Similar taxonomic structures have been reported in marine and freshwater biofilms (Salta et al. 2013, Besemer 2015).
The diversity of bacteria detected in the biofilms indicates the diverse biological functions of these microbial conglomerates and a very complex interaction network. There is evidence demonstrating that mature biofilms are still physiologically active and are not merely aggregates on surfaces (Watnick and Kolter 2000, Vargas-Albores et al. 2019).
Considerable differences were detected between the bacterial profiles of both biofilms; however, they shared 10 of the 27 species observed in all the samples. From these, H. hydrossis seems to play a major role in the biology and physical structure of both biofilms. This species was detected in high proportions in all biofilms and is strictly an aerobic (gram-negative), filamentous, biomass bulking bacterium (Kotay et al. 2011), which grows in strands or filaments rather than a floc; these filaments are reported to be abundant in aquatic systems with high surface:volume ratios (Kotay et al. 2011). In addition, this and other phylogenetically close bacteria have been reported to produce adhesin-like compounds (Larsen et al. 2008), which may enable tight attachments to submerged surfaces.
The high relative abundance of K. vulgare in phototrophic and heterotrophic biofilms suggests that the species plays an important role in the biology of these biofilms. This species is gram-negative, facultative anaerobic, and non-motile and can use a wide spectrum of carbon sources, including several exopolymeric substances produced by biofilms (Staats et al. 1999). It has been recognized as a producer of vitamin precursors, especially 2-keto-L-gulonic acid (vitamin C precursor), which can be used by other bacteria or microalgae. Ketogulonicigenium vulgare can also interact with diverse bacterial groups to form complex interaction networks, as occurs in biofilms (Cai et al. 2012).
Planctomyces (Planctomycetes) was another aerobic, chemoorganotrophic genus detected in both biofilms; this genus has been reported to be present in marine environments and its detection in the biofilms suggests a role in the decomposition of nitrogenous waste (Kuypers et al. 2003). Moreover, other bacteria known to be metabolizers of nitrogenous and sulfurous compounds were exclusively detected in each type of biofilm. Despite the metabolic activity not being monitored in this study, these results could be considered evidence of the potential metabolic capabilities of these microbial conglomerates.
As expected, autotrophic bacteria were also detected in the PAb; for instance, the detection of ammonia-oxidizing bacterium N. multiformis suggests participation in the nitrogen cycle of the system; previous reports have shown that members of the Nitrosospira genus are important nitrogen recyclers in aquatic ecosystems (Okabe et al. 2004, Gomez-Villalba et al. 2006). Furthermore, the presence of abundant gene clusters encoding proteins associated with exopolysaccharide synthesis have been reported for N. multiformis (Norton et al. 2008), and synthesized exopolysaccharides may serve as an energy source for other heterotrophic bacteria. Heterotrophic bacteria were also detected in this biofilm; for instance, the exclusive presence of heterotrophic R. baltica in PAb indicates a biological role of the biofilm in the recycling of sulfurous compounds. This species is a marine, gram positive bacterium that is globally distributed and environmentally important, containing 110 genes coding for sulfatases (Glöckner et al. 2003), which play significant roles in the cycling of sulfur in the environment. However, the genome sequences of many Planctomycetes have a very large repertoire of putative sulfatases, whose role is still unknown.
The candidate division WWE3 bacterium (Waste Water of Evry 3), exclusively detected in the PAb, has been previously isolated from freshwater biofilms, inside sludge flocs, and in anaerobic digesters. However, fosmid annotations have not been conclusive regarding any particular metabolic activity (Guermazi et al. 2008), but their continuous detection in bacterial conglomerates suggests a possible role in the complex interaction network formed within phototrophic biofilms. Echinicola vietnamensis was also exclusively detected in PAb, but its role is also unclear because of the scarce scientific information regarding its biology. Echinicola vietnamensis is a heterotrophic, gram-negative bacterium that was first detected in 2007 in a mussel farm (Nedashkovskaya et al. 2007).
An interesting result is the exclusive presence of S. grandis in PAb; this is a gram-negative, marine, multicellular, chemoorganotroph, filamentous-flexibacterium (Delk and Dekker 1972, Lewin 1997). It could play a role in the decomposition of organic material, and also act as a predator of other bacteria and algae, thereby controlling the population and preventing excessive bacterial/algal blooms. Predator bacteria were also detected in the heterotrophic biofilm. For instance, M. aeruginosavorus was only detected in the Hb; this is an obligate predator that attaches to the surface of a broad range of gram-negative bacteria and feeds by leaching (Kadouri et al. 2007, Dashiff et al. 2011). It has previously only been detected in wastewaters, and this is the first time it has been detected in biofilms and aquaculture systems. The presence of several gram-negative bacteria, such as Arcobacter sp. L. and M. testaceum may provide optimal conditions for the proliferation of gram-negative predators. However, the presence of M. testaceum in the heterotrophic biofilm was unexpected considering that this gram-negative species has been only detected in terrestrial plants (Morohoshi et al. 2011).
More marine heterotrophic species were detected in these biofilms, for instance, the ubiquitous H. neptunium involved in nitrogen recycling processes (Badger et al. 2006). The Hyphomonas genus uses a wide range of substrates, including nitrogen, mainly from proteins and amino acids, as the major energy source of its metabolism. The presence of bacteria containing genes for the metabolism of nitrogenous and sulfurous compounds is of paramount importance for aquaculturing. The results indicate that other bacteria, apart from those already known, could also play significant roles in the cycle of these nutrients, either organic or inorganic.
Animal- and human-associated pathogens such as Parachlamydia acanthamoebae, S. negevensis, and S. agalactiae were detected in both biofilms, while others were exclusive to a particular biofilm type. For instance, Arcobacter sp. and R. anatipestifer were exclusive to the Hb. Simkania negevensis is an amoebal apparent-obligate endoparasite unable to be cultured on conventional laboratory media (Kahane et al. 2001) and S. agalactiae strains are gram positive, facultative anaerobic bacteria that have been isolated from the marine environment, including from marine mammals and fish (Evans et al. 2008). Thus, it is possible that water is a vector of these bacteria and biofilms provided the optimal conditions for their proliferation. We have previously reported for the first time all of these species in farmed shrimp (Vargas-Albores et al. 2017), but their role, along with those of others, in biofilm and shrimp biology remains unclear.
The diversity of bacteria found in these biofilms suggests a complex microbial community and reveals the wide bacterial diversity of microbial conglomerates. Moreover, less than 50% of the reads were taxonomically classified up to species level, which suggests that the diversity in a single bacterial biofilm could be estimated at the level of hundreds. In addition, the microbial network of microbes is even more complex considering that microalgae, fungus, and other microbes are also important constituents of the biofilm. For instance, benthic microalgae, such as Navicula sp. and others, can form biofilms and produce exopolymeric substances that may serve as energy sources for heterotrophic bacteria (Bruckner et al. 2008).
It is worth noting that most of the bacteria detected in both biofilms were first detected in the water used to culture these microbes; however, the different conditions and biology of each consortium caused the growth of specific taxa which may provide functions required under the specific conditions.
The relative abundance of filamentous bacteria in PAb was lower than that of the Hb, most likely due to the fact that the previous colonization of the substrate by the benthic diatom Navicula sp. provided advantages to the microalga over the bacteria; on the other hand, the Hb depended more on the biofouling capacity of filamentous bacteria.
It is difficult to discuss some of the bacteria present in the biofilms in great detail since they have either been recently discovered or are bacteria identified for the first time in biofilms and aquaculture systems. Further functional potential analyses may provide additional information. However, the present study highlights the importance of bacterial characterization in shrimp aquaculture biofilms and provides an insight into the biology of these microbial conglomerates, which are currently revolutionizing aquaculturing.











 nova página do texto(beta)
nova página do texto(beta)


