Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.37 no.3 Texcoco sep. 2019 Epub 30-Sep-2020
https://doi.org/10.18781/r.mex.fit.1905-3
Phytopathological notes
Phyllody of daisy (Dimorphotheca sinuata) associated to Candidatus phytoplasma asteris
1 Postgrado en Fitosanidad-Fitopatología, Colegio de Postgraduados, Carretera México-Texcoco Km 36.5, Montecillo, Texcoco 56230, Estado de México, México.
2 Herbario JES, Área de Biología, Departamento de Preparatoria Agrícola, Universidad Autónoma Chapingo. Texcoco, 56230, Estado de México.
In Montecillo Texcoco, State of Mexico we observed daisy plants (Dimorphotheca sinuata) showing symptoms of phyllody, virescence and proliferation of axillary branches, all those induced by phytoplasmas. Therefore, the objective of this investigation was to detect the phytoplasma associated with these symptoms by PCR with primers P1/P7 and R16F2n / R2 (nested PCR). The 1200 bp PCR product was obtained from the PCR and it was digested with the restriction enzymes MseI (Tru91), AluI, Kpn1 and HaIII and restriction maps showed that the phytoplasma present in Dimorphoteca sinuata is Candidatus phytoplasma asteris (16Sr1-B). The sequences were deposited on the NCBI data base and had a similarity of 99% with Candidatus phytoplasma asteris group 16Sr1-B from Iran (MH638316.1). A phylogenetic analysis was performed with the Neighbour-Joining method in which the phytoplasm from daisy was grouped with Candidatus phytoplasma asteris. According to the symptoms observed in the field, analysis of restriction patterns, sequencing and phylogeny, the phyllody of daisy is associated with Candidatus phytoplasma asteris, phylogenetically related to the 16Sr1-B group.
Key words: Floral reversion; RFLP; sequencing; phytoplasms
En Montecillo Texcoco, Estado de México se observaron plantas de margarita (Dimorphotheca sinuata) exhibiendo síntomas de filodia, virescencia y proliferación de ramas axilares putativos a los inducidos por fitoplasmas. Por lo que el objetivo de esta investigación fue detectar el fitoplasma asociado a estos síntomas mediante PCR con los iniciadores P1/P7 y R16F2n/R2. Se detectó la presencia de fitoplasmas en tejido foliar sintomático. El producto de PCR de 1200 pb obtenido de la PCR se secuenció y sometió a digestión con las enzimas de restricción MseI (Tru91), AluI, Kpn1 y HaIII, los patrones de restricción evidenciaron que el fitoplasma presente en Dimorphotheca sinuata es Candidatus phytoplasma asteris (16SrI). Las secuencias obtenidas fueron depositadas en la base de datos del NCBI y tuvieron una similitud del 99% con Candidatus phytoplasm asteris grupo 16Sr1-B de Irán (MH638316.1). Se realizó un análisis filogenético con el método de Neighbour-Joining, en el cual el aislamiento detectado en margarita, se agrupó con Candidatus phytoplasma asteris. De acuerdo con los síntomas observados en campo, análisis de patrones de restricción, secuenciación y filogenia, indican que la filodia en margarita está asociada a Candidatus phytoplasma asteris, relacionado filogenéticamente al grupo 16Sr1-B.
Palabras clave: Reversión floral; RFLP; secuenciación; fitoplasmas
Phytoplasmas are non-cultivable bacteria with no cell walls, belonging to the class Mollicutes (Weisburg et al., 1989), and are responsible for hundreds of diseases in cultivated plants and weeds around the world (Lee and Gundersen-Rindal, 2000). These pathogens are restricted to the phloem, are transmitted by grafting and by insects (Weintraub and Beanland, 2006), as well as by seed (Rojas-Matínez et al., 2009). Phytoplasmas systematically infect their hosts by moving through the pores of the plates of the phloem, distributing throughout their vascular system (Lee and Gundersen-Rindal, 2000). So far, these microorganisms are not cultivated in a free cell medium, which suggests that they have a more reduced metabolism than other mollicutes, as observed in the genomes of phytoplasmas that have been sequenced to date (Oshima et al., 2004).
Phytoplasmas induce a variety of symptoms, including yellowing, delay in growth, sterility of flowers, necrosis, witche’s broom, phyllody and virescence, among others (Rojas et al., 2013). The three latter symptoms suggest that phytoplasmas interfere with the metabolism of plant hormones (Weintraub and Bealand, 2006). The severity of symptoms depends of the phytoplasma isolate, the age of the plant and the moment in which the infection occurs.
The interaction of phytoplasmas with their vector insects is complex and implies its intra- and extra-cellular replication in the intestine, salivary glands, epithelial and muscular tissues and other organs. The systemic infection of the phytoplasms inside the insect may take ten days or more, depending on the group, the species of the insect, and temperature (Sugio and Hogenhout, 2012). Considering that there is evidence that some phytoplasmas are transmitted vertically in their vector insects (Weintraub and Bealand, 2006), the most effective means of survival is by this way. The insects that can be vectors of these pathogens belong mainly to the families of Cicadellidae, Fulgoridae and Psilidae in smaller numbers. (Weintraub and Bealand, 2006).
So far there are four phytoplasma genomes completely sequenced, including the causal agent of Aster yellows (Candidatus Phytoplasma asteris), which has a wide range of hosts and is transmitted by different polyphagous insects. Several factors contribute to the reduction of the phytoplasma genome, including small population sizes, asexuality, a mutational bias that favors eliminations over insertions, an environment metabolically rich of growth, and finally, the absence of a flow of the genome from other sources, due both to the restricted intracellular environment as well as the inability to incorporate foreign DNA by recombination (Bai et al., 2006). Although the phytoplasmas are subjected to most of these factors, they are not restricted to a single insect, and populations can consist of multiple phytoplasma variants (Weintraub and Bealand 2006). In particular, the phytoplasmas that colonize many plants and insects are more prone to find other phytoplasmas and organisms that constitute sources for the acquisition of genetic elements.
Due to the economic importance of these pathogens and to the scarce knowledge on vector insect species in Mexico, it is necessary to know weeds and/or ornamental plants with the potential of becoming reservoirs of these pathogens and contribute to the knowledge on them. Therefore, the aim of this investigation was to identify the phytoplasma that induces phyllody in daisy plants (Dimorphotheca sinuata).
DNA extraction. DNA was extracted from leaves in five plants with symptoms of phyllody, (Figure 1); as well as from two asymptomatic plants as a control. The method used for the extraction of DNA was the reported by Dellaporta et al. (1983) with some modifications.
Detection of phytoplasmas by PCR. Nested PCR was carried out using universal primers P1 (Deng and Hiruki, 1991) and P7 (Kirkpatrick et al., 1994) for phytoplasmas that amplify a fragment of 1800 pb (first amplification) in a final volume of 25 µL that contained 1 X of PCR buffer (10 X, 100 mM tris-HCl, 500 mM KCl, pH 8.3) (Invitrogen®), 0.2 mM of every dNTP, 1.5 mM of MgCl2 (Invitrogen®), 10 pmol of each primer (Sigma-Aldrich®), 1 U of DNA (Invitrogen®) and 100 ng of DNA template. The amplification polymerase was carried out in a Techne® thermocycler TC-300 with the following program: denaturalization at 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, 55 °C for 2 min, 72 °C for 3 min with a final extension of 72 °C for 5 min. The second amplification (nested) was carried out using primers R16F2n / R16R2 (Gundersen and Lee, 1996) that amplify a fragment of 1200 pb of the region 16S rADN of the phytoplasmas. As a template DNA, we used the amplified product of the first reaction of PCR diluted in sterile water, free of nucleases (1:20) using the same concentrations and reactants than in direct amplification, with the following program: denaturalization at 94 °C for 2 min, followed by 35 cycles at 94 °C for 1 min, 58 °C for 2 min, 72 °C for 3 min and a final extension at 72 °C for 10 min.
Sequencing and phylogenetic analysis. The product obtained from PCR was purified and sequenced (Macrogen Inc. Korea) in both directions. The sequences obtained were analyzed, deposited and compared with those included in the National Center for Biotechnology Information (NCBI, 2016) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the BLAST. With the sequence obtained, we carried out a tool phylogenetic analysis with other species of phytoplasmas (Figure 3).

Figure 1 A-C. Daisy plants showing symptoms of phyllody and proliferation of sprouts, (D) asymptomatic plants.
In all samples with symptoms, we obtained the expected fragment of 1200 pb for phytoplasmas (Figura 2A). The sequences obtained (Access No. MK278895 and MK278896) had a similarity of 99% with Candidatus Phytoplasma asteris group 16srI-B obtained from Vitis vinifera in Iran (MH638316.1). The analysis of RFLP in vitro (Figure 2B) indicated that the phytoplasm of this study belongs to the group of the Aster yellow, now recognized as Candidatus Phytoplasma asteris.
The use of restriction enzymes to identify groups of phytoplasmas are still useful, since the electrophoretic profiles generated by each of the enzymes are an indicator of the variation present in their genome, and therefore these patterns become a genetic footprint. In many investigations, the enzymes most used for this purpose are MseI (Tru 91), AluI, RsaI, Hha, HpaII and HpaIII (De Oliveira et al., 2011). The phytoplasmas of the Aster yellow constitute a cosmopolitan group that is found in most plants in natural conditions, due to its plasticity to adapt to many hosts, and they can therefore eventually become a serious problem (Lee et al., 2000). Most symptoms induced by phytoplasmas are known to be the same in different species, and that a crop or a single plant may contain more than one phytoplasma. Because the symptomatology is the same, one may ignore the infectious potential of some may have and disseminate to crops of agronomical interest, leading to a possible economic problem. In Mexico, most studies are focused on detecting these pathogens, and in a few cases, such as the lethal yellowing of palm, work has been done with genetic breeding and replacements of material in areas seriously affected by the disease. Another disease of economic impact in a field caused by a phytoplasma is the thickening of the cladodes of prickly pear, since it reduces considerably the production of prickly pears in all production areas (Suaste et al., 2012). In recent years, the presence of phytoplasmas has been observed in ornamental plants introduced into Mexico (Rojas et al., 2017), since they showing abundant proliferations, and in some cases, green flowers that consumers find very attractive; this situation may favor the appearance of emerging diseases. On the other hand, the literature indicates that symptoms may appear one week after the inoculation of the phytoplasma; however, this depends on the temperature conditions and the plant species. The severity of the symptoms varies according to the age of the plant, the isolate of phytoplasma and the time of infection (Weintraub and Bealand 2006). Likewise, the phytoplasma is occasionally detected in symptomatic plants that presented no alteration in its period of development (Rojas et al., 1999). Some phytoplasmas are acquired and transmitted exclusively by one species of insects; however, others are transmitted by different insect species. From an epidemiological point of view, the transmission of phytoplasmas by insects is particularly important if they are polyphagous, as in the case of cicadellids, which also constitute an important reservoir of these pathogens (Weintraub and Bealand 2006).

Figure 2 (A) Products of PCR obtained with primers R16F2n/R16R2. Agarose gel at 1%. Lanes 1-5, samples of daisies with symptoms of phyllody; Lanes 6 y 7, asymptomatic samples; Lane 8, Negative control (water); Lane 1kb, molecu lar marker 1kb (Promega®). (B) Digestion of the PCR products with restriction enzymes. Lanes 1 and 10 marker 1kb (Promega®), Lanes 2 and 3 product of digestion with AluI; Lanes 4 and 5 digestion with Kpn1; Lanes 6 and 7 product of the digestion with Tru9I; Lanes 8 and 9 products of the digestion with Hae III.
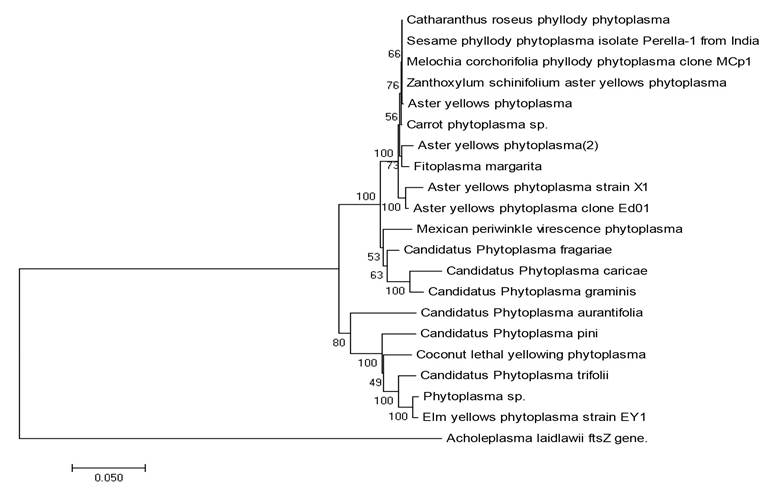
Figure 3 Phylogenetic tree based on ribosomal DNA 16S of phytoplasma sequences available in the NCBI. Acholeplasma laidlawii, a that can be cultivated in vitro and that is not genetically related to phytoplasmas data base, was consid ered as a root. The tree was created using p CLUSTAL_X with the neighbor-joining method with 500 repetitions.
Phytoplasmas have a metabolically limited genome with moving elements that contain information to codify different effectors that are transcription factors that modulate diverse responses in the plant (Bai et al., 2006). As a result of this, alterations such as witche’s broom are produced, as well as changes in the structure and color of the leaves or suppression in the defense response of the plant to vector insects of these pathogens (Cettul and Fierrao, 2011). Finally, it is worth investigating insects that colonize this ornamental plant, since it is widely distributed in Mexico, particularly adjacent to peach, plum, cactus, broadbean, bean and pumpkin crops, among others.
With the detection of Dimorphotheca sinuata as a new host of Candidatus Phytoplasma asteris, the knowledge on the range of alternate hosts for this group of phytoplasmas broadens, and turns it into a potential source of inoculant for other crops of economic interest that host this phytoplasma, such as oat, where the subgroup 16SrI-B induces serious damage to this crop consisting in an abnormal proliferation and yellowing and sterilization of spikes, delay in growth and the production of sterile seeds (Urbanavičienė et al., 2006). In this ornamental species (Dimorphotheca sinuata), one phytoplasma belonging to group 16SrIX, causing phyllody, growth delay and virescence in Italy (Marcone et al., 2001) had been reported.
The symptoms of phyllody and proliferation of daisy (Dimorphotheca sinuata) were associated with Candidatus Phytoplasma asteris, phylogenetically related to group 16Sr1-B. Dimorphotheca sinuata is reported, for the first time, as a new host for Candidatus Phytoplasma asteris.
Literatura citada
Bai X, Zhang J, Ewing A, Miller SA, Jancso RA, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A and Campbell JW. 2006 Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. Journal of Bacteriology 188:3682-3696. DOI:10.1128/JB.188.10.3682-3696. [ Links ]
Cettul E and Firrao G. 2011. Development of phytoplasma-induced flower symptoms in Arabidopsis thaliana. Physiology and Molecular Plant Pathology 76:204-211. DOI:10.1016/j.pmpp.2011.09.001 [ Links ]
De Oliveira AP, Mello A, Eckstein B, Flores D, Fabretti KP and Bedendo IP. 2011. Identification by computer-simulated RFLP of phytoplasmas associated with eggplant giant calyx representative of two subgroups, a lineage of 16SrIII-J and the new subgroup 16SrIII-U. International Journal of Systematic and Evolutionary Microbiology. 61: 1454-1461. DOI: 10.1099/ijs.0.019141-0 [ Links ]
Deng, SJ and Hiruki, C. 1991. Genetic relatedness between to non culturable mycoplasmalike organisms revealed by nucleic acid hybridization and polimerase chain reaction. Phytopatology 81: 1475-1479. DOI: 10.1094/Phyto-81-1475 [ Links ]
Dellaporta SL, Wood J and Hicks JB. 1983. A plant DNA Minipreparation: Version II. Plant Molecular Biology Report. 1:19-21. DOI: 10.1007/BF02712670 [ Links ]
Gundersen DE and Lee I-M. 1996. Ultrasensitive detection of phytoplasmas by nested- PCR assays using two universal primer pairs. Phytopathology Mediterranea. 35:114-51. https://www.jstor.org/stable/42685262 [ Links ]
Hogenhout, SA and Segura-Music, M. 2010. Phytoplasma genomics, from sequencing to comparative and functional genomics-What have we learnt? pág. 19-36. eds P.G. Weintraub and P. Jones In: Phytoplasmas: Genomes, Plant Hosts and Vectors http://ebookcentral.proquest.com/lib/colpos-ebooks/detail.action?docID=476509 [ Links ]
Kirkpatrick B, Smart C, Gardner S, Gao JL, Ahrens U, Mäurer R, Schneider B, Lorenz, K‐H, Seemüller E, Harrison N, Namba S and Daire X. 1994. Phylogenetic relationships of plant pathogenic MLOs established by 16/23S rDNA spacer sequences. IOM Letters 3, 228- 9. [ Links ]
Lee IM, Davis RE and Gundersen-Rindal DE. 2000. Phytoplasma: phytopathogenic mollicutes. Annual Review of Microbiology 54:221-255. DOI:org/10.1146/annurev.micro. [ Links ]
Marcone C, Ragozzino A, Camele I, Rana GL, Seemüller E. 2001. Updating and extending genetic characterization and classification of phytoplasmas from wild and cultivated plants in southern Italy. Journal of Plant Pathology 83, 133-138. [ Links ]
Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S and Ugaki M. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nature Genetics 36:27-29. DOI: 10.1038/ng1277 [ Links ]
Rojas-Martínez RI, Zavaleta-Mejía E, Trejo LC, and Azpiroz HR. 1999. Some physiologycal alterations in marigold (Tagetes erecta L.) plants infected by phytoplasmas. Fitopatología. 34:225--229.https://www.researchgate.net/profile/Emma_Zavaleta-Mejia/publication/295073546_Some_physiological_alterations_in_marigold_Tagetes_erecta_L_plants_infected_by_phytoplasmas/links/56c6d15708ae0d3b1b6179eb/Some-physiological-alterations-in-marigold-Tagetes-erecta-L-plants-infected-by-phytoplasmas.pdf?origin=publication_list [ Links ]
Rojas-Martínez RI, Ochoa Martínez D L y Zavaleta Mejía E. 2013. Fitoplasmas y Ca. Liberibacter sp. en cultivos agrícolas. Colegio de Postgraduados, Montecillo, Texcoco, Edo de México. 56p. ISBN 978-607-715-138-8. [ Links ]
Rojas-Martínez RI, Zavaleta-Mejía E e Ing-Ming L. 2009. Identificación de un aislamiento del grupo 16SRIII Candidatus Phytoplasma pruni en plantas y semillas de amaranto (Amaranthus hypochondriacus l.) en México. Agrociencia. 43:851-860. [ Links ]
Suaste DA, Rojas MRI, Zavaleta ME y Pérez BD. 2012. Detección molecular de fitoplasmas en nopal tunero (Opuntia ficus-indica) con síntomas de engrosamiento del cladodio. Revista Mexicana de Fitopatología 30:72-80. http://www.redalyc.org/articulo.oa?id=61225129007 [ Links ]
Sugio A and Hogenhout SA. 2012. The genome biology of phytoplasma: modulator of planst and insects. Current Opinion in Microbiology, 15:247-254. DOI:10.1016/j.mib.2012.04.002: [ Links ]
Weisburg WG, Tully JG, Rose DL,Petzel JP, Oyaizu H,YangD, Mandelco L, Sechrest J, Lawrence TG and VanEtten J. 1989. A phylogenetic analysis of the mycoplasmas:basis for their classification. Journal of Bacteriology. 171:6455-67. DOI:10.1128/jb.171.12.6455-6467.1989. [ Links ]
Weintraub P and Bealand A. 2006. Insect vectors of phytoplasmas. Annual Review of Entomology 51: 91-111. DOI:10.1146/annurev.ento.51.110104.151039. [ Links ]
Urbanavičienė L, Jomantienė R, Valiūnas D, Davis RE. 2007. Molecular identification of 16SrIA16SrI-B, 16SrI-C, and 16SrI-L subgroups of phytoplasmas in gramineous plants in Lithuania. Bulletin of Insectology 60, 127-128. [ Links ]
Marcone C, Ragozzino A, Camele I, Rana GL, Seemüller E. 2001. Updating and extending genetic characterization and classification of phytoplasmas from wild and cultivated plants in southern Italy. Journal of Plant Pathology 83, 133-138. [ Links ]
Received: May 17, 2019; Accepted: July 31, 2019











 texto en
texto en 


