Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.37 no.3 Texcoco sep. 2019 Epub 30-Sep-2020
https://doi.org/10.18781/r.mex.fit.1905-5
Scientific articles
PCR detection of Guignardia bidwellii, causal agent of grape black rot
1 Centro Nacional de Referencia Fitosanitaria, SENASICA SAGARPA, Km 37.5 Carretera Federal México-Pachuca, Tecámac Estado de México C. P. 55740.
Guignardia bidwellii (anamorph Phyllosticta ampelicida) is the etiological agent of grape black rot, a disease of economic importance in Europe and a quarantine pest for Mexico. The identification of the anamorphic state by morphological characteristics is complicated, due to the similarity among species of the same genus, for example with the cosmopolitan endophyte G. endophyllicola (P. capitalensis), therefore it is necessary to have quick and specific diagnostic tools. For this purpose, a molecular marker based on single nucleotide polymorphisms (SNP) in the ITS region of the rDNA was developed, where the Bidwell and Ampel2 primers were designed. The PCR amplification generates a fragment of 173 bp, specific to G. bidwellii f. euvitis. The validation of the method showed that there is no cross amplification with closely related phytopathogenic fungi or with the genome of grapevine. The technique was sensitive to detect up to 30 pg μL-1 of DNA from monosporic cultures and in mixture of plant tissue. This diagnostic procedure is presented as a fast and specific option for the monitoring, and detection of G. bidwellii f. euvitis in support of prevention, management, quarantine and eradication strategies of the disease.
Key words: Phyllosticta ampelicida; molecular diagnosis; ITS; Vitis spp
Guignardia bidwellii (anamorfo: Phyllosticta ampelicida) es el agente etiológico de la pudrición negra de la vid, enfermedad de importancia económica en Europa y cuarentenaria para México. La identificación del estado anamorfo mediante caracteres morfológicos resulta complicada debido a la similitud entre especies del mismo género, por ejemplo, con el endófito cosmopolita G. endophyllicola (P. capitalensis), por lo que es necesario contar con herramientas de diagnóstico rápidas y específicas. Con este propósito se desarrolló un marcador molecular basado en los polimorfismos de un solo nucleótido (SNP) en la región ITS del ADNr, donde se diseñaron los oligos Bidwell y Ampel2 que mediante amplificación PCR generan un fragmento de 173 pb específico a G. bidwellii f. euvitis. La validación del método demostró que no hay amplificación cruzada con otros hongos fitopatógenos ni con el genoma de la vid. La técnica fue sensible al detectar hasta 30 pg µL-1 de ADN a partir de cultivos monospóricos y en mezcla con tejido vegetal. Se presenta este procedimiento de diagnóstico como una opción rápida y específica para el monitoreo y detección de G. bidwellii f. euvitis en apoyo a las estrategias de prevención, manejo, cuarentena y erradicación de la enfermedad.
Palabras clave: Phyllosticta ampelicida¸ diagnóstico molecular; ITS; Vitis spp
The Guignardia bidwellii phytopathogen fungus (anamorph: Phyllosticta ampelicida) is the causal agent of grape black rot, one of the most important diseases that affect grape production in Europe (Ullrich et al., 2009). The fungus is native to North America and was introduced into Europe when infected hybrids were mobilized at the end of the XIX century (Wicht et al., 2012). The pathogen is distributed across important grape producing zones such as France, Germany and northern Italy, a fact that poses a potential phytosanitary risk in those regions, where relative humidity is high in the summer and causes considerable economic losses if there are no adequate measures for controlling the disease (Wicht et al., 2014).
Black rot affects mainly grape crops (Vitis spp.) but is also present in other hosts of the Vitaceae family: Parthenocissus spp., Ampelopsis spp., Cissus spp. (Van der Aa, 1973; Wicht et al., 2012). Differences have been observed among diverse cultivars, but all the commercially important varieties are susceptible to the disease (Ullrich et al., 2009). Luttrell (1946) proposed that the fungus has three variants or forms (f.) depending on the host (f. euvitis, f. parthenocissi and f. muscadinii). However, molecular studies have demonstrated differentiation only in two different forms: those coming from Vitis and Parthenocissus hosts (Wicht et al., 2014). Traditional identification of G. bidwellii is based on the description of morphological traits and symptoms expression (Wicht et al., 2012), but the anamorphic state has characteristics similar to several related-species, for example, pycnidia with non-septate, hyaline conidia covered by a mucoid layer, and an apical appendix (Wikee et al., 2013a).
The Guignardia endophyllicola (Phyllosticta capitalensis) endophyte fungus is a cosmopolitan species and a weak parasite in a wide range of hosts (Okane et al., 2003) which includes members of the Vitaceae family (Wikee et al., 2013b). The telemorphic and anamorphic states can be morphologically confounded with G. bidwellii and P. ampelicida, which leads to an incorrect identification, thus being necessary to corroborate the results using molecular techniques (Wicht et al., 2014).
Only a few molecular studies about G. bidwellii have been conducted. In a first approximation, Wicht et al. (2012) performed a phylogenetic analysis of ITS1 and ITS2 regions of samples from diverse geographical origins and concluded that there was evidence of two variants associated with hosts of the Vitis and Parthenocissus genera. Zhang et al., (2013) reached a similar conclusion through a multilocus analysis of four genes and proposed to re-categorize this form at the level species as P. parthenocissi. In the meantime, the use of microsatellites has made possible to determine that there is a great allelic variety in the populations in Europe and between both hosts (Wicht et al., 2014). However, there is not enough evidence to associate a G. bidwellii genotype with one specific reaction in the Vitis host (Rinaldi et al., 2017).
Currently, grape black rot is an economically important disease in Europe and a quarantine pest for Mexico (SENASICA, 2019), so as part of the scientific support to the epidemiological surveillance programs, it is important to have a protocol based on molecular techniques that allow a correct identification of the species. Therefore, the objective of the present study was to design, develop and validate a molecular marker based on the PCR technique using specific oligos to detect G. bidwellii.
Materials and methods
Obtaining isolates. For the experiment, Guignardia bidwellii f. euvitis ATCC 9560 (Georgia, United States), Guignardia endophyllicola CNRF-MICO-C1/001 (Michoacán, Mexico) and Guignardia citricarpa ATCC 26254 (South Africa) were used as reference strains, as well as isolates of phytopathogen species Verticillium dahliae, Fusarium oxysporum, Alternaria alternata, Colletotrichum gloeosporioides from the phytopathogen fungi collection of the National Phytosanitary Reference Center (CNRF, for its acronym in Spanish) of the General Directorate of Plant Health (DGSV, for its acronym in Spanish) in Mexico, and grape leaves infected with Plasmopara viticola.
The isolates of the CNRF-SENASICA collection are kept in mineral oil at 4 °C, according to the methodology used by Humber (1997) and were reactivated in a potato-dextrose-agar (PDA) medium and incubated at 25 °C ± 3 °C for 10 days. Then, monosporic cultures, monopycnidial cultures in the case of gender Guignardia, were carried out in PDA, based on Crous et al., (2005), and incubated at the previously mentioned temperature under 12/12 hours light-darkness intervals for 15 days. P. viticola was preserved in the form of herbarium in a botanical press.
DNA extraction. Total genomic DNA was extracted from species of the Guignardia genus and from grape tissue, healthy and infected with P. viticola, following the method of CTAB (Doyle and Doyle, 1987); for the other four species, DNA was extracted according to the methodology proposed by Cenis (1992), that is, using PDA plates but omitting the first centrifugation. The amount and quality of DNA was verified through the absorbance relation A260/280 and A260/230 (Manchester, 1995) in a Nanocrop 2000C spectrophotometer.
Design of oligos. For in silico tests, sequences of ITS regions of rDNA of 46 species of the Guignardia genus reported by Wikee et al. (2013a) were used, as well as the reference sequences AB095505, AB095509 to AB095511, AB454268, AB454276, AB454313, HM008727 to HM008728, FJ824766, EU683672, KC193586, KF015253 to KF015268 and KF851288 to KF851317 corresponding to f. euvitis and f. parthenocissus (Okane et al., 2003, Motohashi et al., 2009, Wicht et al., 2012; Zhang et al., 2013; Rinaldi et al., 2017). The sequences were aligned using the ClustalW algorithm in BioEdit 7.2.5 (Hall, 1999). Single nucleotide polymorphisms (SNP) characteristic to G. bidwellii f. euvitis consensus reference were identified with respect to the other 46 species, and the ITS region was used to design the forward Bidwell oligonucleotide.
The reverse Ampel2 oligonucleotide was designed with the Primer3 software (Untergasser et al., 2012) using the same consensus sequence. The pair of oligonucleotides was structured in order to amplify a unique 173 bp fragment. The thermodynamic parameters, the formation of internal structures, the heterodimers and the autodimers were examined in silico using mFold (Zuker, 2003). The specificity of both oligonucleotides was tested in silico with Primer BLAST (Ye et al., 2012).
PCR amplification. After extraction, a PCR test of the endogenous gene was conducted using ITS1 oligonucleotides (5´ TCCGTAGGTGAACCTGCGG 3´) and ITS4 (5´ TCCTCCGCTTATTGATATGC 3´) (White et al., 1990) to verify the quality of the genetic material. The amplification was performed with a Taq DNA Polymerase kit from Invitrogen®, the reaction mixture was prepared using a final concentration of Buffer 1X, 1.5 mM of MgCl2, 0.2 mM of dNTPs, 0.5 µM of each primer, 2.5 U Taq Polymerase and 2 ng µL-1 of DNA in a 25 µL volume. The thermal program used was an initial denaturation at 95 °C for 3 min, followed by 35 cycles at 95 °C for 45 s, hybridization at 58 °C for 45 s, an extension at 72 °C for 45 s and a final extension at 72 °C for 10 min. For this, a T100™ Thermal Cycler from BioRad was used.
The amplification of the 173 bp fragment using the specific oligonucleotides was performed using the same reaction mixture with a thermal program of denaturation at 95 °C for 5 min, followed by 35 cycles at 95 °C for 45 s, hybridization at 62 °C for 30 s, one extension at 72 °C for 30 s and a final extension at 72 °C for 5 min. The PCR products were analyzed in 2% ultra-pure agarose gels in a TAE 1X buffer marked with 1X from GelRed® Biotium. The size of the amplified fragments was estimated using a TrackIt 100 bp DNA Ladder marker from Invitrogen.
Positive control. The positive control was obtained using the specific fragment of the amplified gene with Bidwell and Ampel2 oligonucleotides. The PCR product was cloned using pGEM® T Easy Vector from Promega. The plasmid carrying the fragment was used to transform One Shot® Mach1™ T1R E. coli competent cells from Invitrogen. The bacterial colonies that were able to introduce the plasmid were selected and DNA extraction was carried out following the alkaline lysis method (Sambrook and Russell, 2001). Then, the cloned insert was amplified by PCR and sequenced in both directions with an ABI PRISM 3130 sequencer from Applied Biosystems® through BigDye™ Terminator v3.1 chemistry.
The sequences obtained were compared to sequences in the database of the National Center for Biotechnology Information (NCBI) using the algorithm known as Basic Local Alignment Search Tool (nucleotide BLAST) (Altschul et al., 1990) and with the sequences reported by Wicht et al. (2012) and Rinaldi et al. (2017).
Specificity test. The pair of oligonucleotides was evaluated against fungi G. bidwellii f. euvitis, G. citricarpa, V. dahliae, F. oxysporum, A. alternata, C. gloeosporioides, P. viticola and G. endophyllicola, from which the latter has been reported as a cosmopolitan fungal endophyte (Okane et al., 2003) morphologically and phylogenetically related to G. bidwellii. DNA of healthy grape tissue was used as a negative control, and one blank control (without any type of DNA). The test was conducted by duplicate including two replications per specimen each time. The result was considered positive when a 173 bp amplified fragment was obtained.
Sensitivity test. Serial dilutions to obtain eight working concentration (3000, 300, 30, 3, 0.3, 0.15, 0.06 and 0.03 ng µL-1) were prepared using a cloned control of G. bidwellii f. euvitis at an initial concentration of 3000 ng µL-1. Each dilution was amplified per triplicate using Bidwell-1 and Ampel-2 oligos under the reaction conditions and the thermal program previously mentioned. Then, a second test was conducted with seven dilutions of the cloned control (1000, 500, 300, 30, 3, 0.3 and 0.03 ng µL-1), two replications per dilution, and 1 µL of DNA extracted from healthy grapes (50 ng µL-1) was added to the 25 µL of the final reaction.
Results and discussion
Design of oligos. The bioinformatic analysis of the oligonucleotides designed for G. bidwellii (Table 1) shows that the value of ΔG was less than 2 kcal mol-1, which indicates that there was no formation of important secondary structures that negatively affect the reaction (Matveeva et al., 2003). Consequently, the values of ΔG for end 3′, autodimers and heterodimers were within the tolerated ranges, according to Untergasser et al. (2012). Other criteria such as fusion temperature (Tm), length, GC %, presence of C or G at end 3´, tandem dinucleotides and repeated bases were within acceptable parameters, according to Rychlik (1995).
Regarding specificity, the analysis in silico of the forward Bidwell oligonucleotide using BLAST showed 100% coverage and identity only with G. bidwellii sequences; no other fungus had an homologous region, and the closest organisms were Streptomyces sp. and Bacillus subtilis with 85% and 76% coverage, respectively. The oligo reverse Ampel2 also showed 100% coverage and identity with G. bidwellii. However, in this case they were completely homologous to many species of Guignardia genus but not to Streptomyces and Bacillus genera.
A multiple alignment with the reference sequences of Wikee et al. (2013a) confirmed that Bidwell is specific, given that the 21 nucleotides of the oligonucleotide are completely homologous to the sequences reported for the Vitis host (Figure 1a). Additionally, the first 11 nucleotides of the oligo end 3′ include the variant reported for the Parthenocissus host (Wicht et al., 2012; Zhang et al., 2013; Rinaldi et al., 2017). On the other hand, Ampel2 is located in a conserved region, since the last eight nucleotides towards end 3´ are constant to all the species of the Guignardia genus (Figure 1b). Both results were in agreement with what we observed in BLAST.
Table 1 Specific oligonucleotides to detect Guignardia bidwellii f. euvitis (Phyllosticta ampelicida) through end-point PCR.
| Oligo | Secuencia 5´a 3´ | pb | Tm (°C)X | %GCX | ΔGy | Valor de Ez | % Coberturaz | % Identidadz |
|---|---|---|---|---|---|---|---|---|
| Bidwell | GAAAAGCCGTCCGAAAGAGCC | 173 | 66.47 | 57.14 | 1.98 | 0.083 | 100 | 100 |
| Ampel2 | CAGGACTTCACGAAATAATCG | 57.38 | 42.86 | 0.84 | 0.083 | 100 | 100 | |
X Values calculated using Primer3 (Untergasser et al., 2012).
y Value of Gibbs free energy calculated using mFOLD (Zuker, 2003).
Z Value of E, coverage and identity determined using nBLAST from NCBI for accessions KF851289 to KF851315.
Amplification and specificity. Genomic DNA of good quality was obtained both for monosporic cultures and for samples from vegetal tissue. The test of the endogenous genes of the ITS region of rDNA had an efficient PCR amplification ranging from 550 to 750 bp according to each species (Figure 2a), which indicates that the DNA obtained is suitable for amplification, is whole and there is no protein inhibition of the reaction.
The PCR reaction with the pair of specific oligonucleotides Bidwell and Ampel2 produced the expected 173 bp fragment only for G. bidwellii DNA (Figure 2b). The oligonucleotides did not show any cross reaction with any other fungus, including G. endophyllicola, which is the most related species at the molecular level. No reaction was either observed in grape tissue, so the plant’s genome does not produce false positives. Oligonucleotides specificity was also confirmed through sequencing of the PCR products that were positive in the test, obtaining a 100% coverage and identity with G. bidwellii accessions KF015253 to KF015255, according to Rinaldi et al. (2017).
Sensitivity test. The amplification of the cloned fragment was inhibited when a concentration of 3000 ng µL-1 was used, so in a second test a concentration of 1000 ng µL-1 was used, where the band was dim or imperceptible. All the other dilutions showed an optimal amplification, which indicates that the method is sensitive because it allows detecting G. bidwellii in concentrations of less than 500 ng µL-1 with a lower limit of detection evaluated in 0.03 ng µL-1. The test with an extract of grape DNA showed similar results, so the sensitivity of the technique was not affected by the presence of the plant’s genome (Figure 3).

Figure 1 Phylogeny of the ITS region of rDNA for the representative Guignardia bidwellii f. euvitis and f. parthenocissi sequences. A) Alignment of the sequences to design forward Bidwell-1. B) Design in direction 5´- 3’ of reverse. Ampel-2. C. gloeosporioides was used as a root node external to the group.

Figure 2 Amplification of the PCR products. A) Test of the endogenous gene with oligos ITS-1 and ITS-4 (550-750 bp). B) Test using specific oligos Bidwell-1 and Ampel-2 (173 bp). For both tests, lane 1-2: G. bidwellii, 3-4: Grape DNA Plasmopara viticola, 5-6: G. endophyllicola, 7-8: G. citricarpa, 9-10: Verticillium dahliae, 11-12: F. oxysporum, 13- 14: A. alternata, 15-16: C. gloeosporioides, 17: Blank with H2O. MM: TrackIt 100 bp DNA Ladder from Invitrogen.
The specific oligonucleotides for G. bidwellii were designed using as reference all the available sequences of G. bidwellii f. euvitis for the ITS region of rDNA, which was chosen to differentiate the species of the Guignardia genus (Wikee et al., 2013a), because it is polymorphic. The internal transcribed spacers of this region have been proposed as a universal DNA Barcoding for the kingdom Fungi because of their high interspecific variability in a wide range of Eumycetos (Schoch et al., 2012). Bonants et al. (2003), Everett and Rees (2006) and Peres et al. (2007) have successfully developed specific oligonucleotides based on the same ITS region of rDNA for other species of the Guignardia genus.
The design of oligos included sequences of representative isolates of the major grape producing regions where the disease is present: United States (Zhang et al., 2013) and Europe (Rinaldi et al., 2017). The sequences of the ATCC 9560 reference control and the cloned control were homologous. Also, for this gene in particular no single-nucleotide polymorphisms were detected for the different geographical origins of the same species, as referred by Wicht et al., (2012). Therefore, although the oligos designed during this study have interspecific strength, the intraspecific variation of subpopulations or haplotypes will require more robust markers (Wicht et al., 2014; Rinaldi et al., 2017).
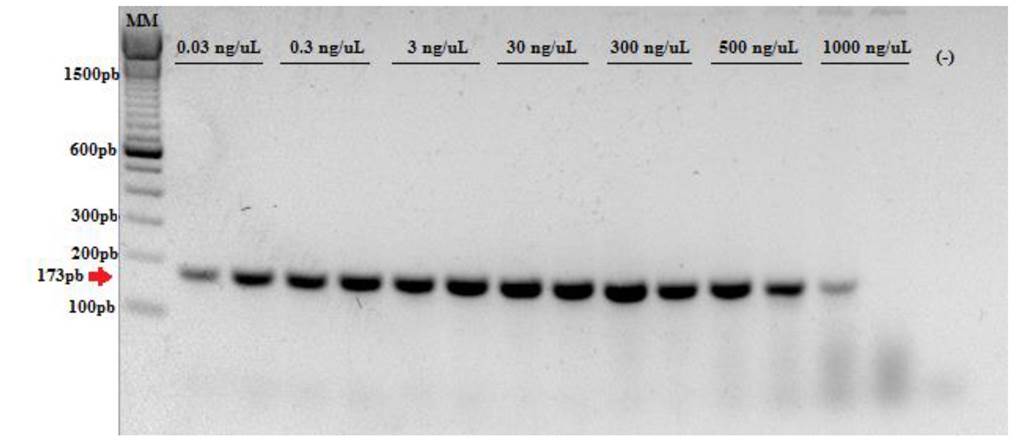
Figure 3 Test of sensitivity of oligonucleotides Bidwell-1 and Ampel-2 using a dilutions gradient of the cloned control in a 50ng μL-1 mixture of grape DNA. (-): Blank with H2O. MM: TrackIt 100 bp DNA Ladder from Invitrogen.
As for the host range and possible specialization or variants cited by Luttrell (1946) and Wicht et al. (2012), the Bidwell and Ampel2 oligos designed are 100% complementary to the sequences reported for the isolates obtained from the host of the Vitis genus; the in silico and in vivo tests produced the expected 173 bp fragment. However, the alignment with the sequences of f. parthenocissi showed an SNP of guanine rather than thymine at position 12 of forward Bidwell, and two additional SNPs at positions 8 and 12 of reverse Ampel2. In in silico tests only one SNP is enough to determine the specificity between two organisms (Stadhouders et al., 2010), but the in vivo specificity could be given only by the first nucleotides of end 3´ (Ayyadevara et al., 2000; Stadhouders et al., 2010). We suggest evaluating oligos for the isolates obtained from Parthenocissus, Ampelopsis and Cissus hosts in future studies.
Considering that, according to Wich et al. (2012), the growth rate of the fungus in culture mediums is slow (fruiting bodies take more than two weeks to form), the protocol described here represents a fast and reliable tool to detect and identify the pathogen, because it allows the use of vegetal material infected or mycelium cultured in vitro that has been growing for less than a week. Similar protocols have been implemented to detect other fungi that are difficult to isolate, such as P. citricarpa (Bonants et al., 2003; Peres et al., 2007) and Elsinoë fawcettii (Hyun et al., 2007). Oligos allow a quick differentiation of G. bidwellii from the rest of species, but do not allow to differentiate a teleomorph from an anamorph, and for this reason they have to be biologically identified. On the other hand, the PCR assay can be complemented by the one proposed by Everett and Rees (2006) for specific detection of the G. endophyllicola endophyte in order to obtain more robust results when diagnosing G. bidwellii f. euvitis (P. ampelicida).
Finally, given that the main source to spread grape black rot is mobilization of propagative material (Wicht et al., 2012), its introduction to grape producing zones poses a great risk for this crop, such as that reported in Europe (Rinaldi et al., 2017). In Mexico, its introduction could affect more than 33 000 ha of grape crops (SIAP, 2018), because there are no resistant cultivars (Ullrich et al., 2009). For this reason, developing fast and accurate diagnosis methods such as PCR must be a priority in order to detect the fungus in time by using propagative vegetal material and implementing adequate confinement and delimitation measures.
Conclusions
This study reports the development of an end-point PCR test based on the ITS region of rDNA to detect and identify in a specific, fast and simple way Guignardia bidwellii f. euvitis (Phyllosticta ampelicida), the causal agent of grape black rot. The test allows to obtain a reliable diagnosis of the fungus and is a starting point for planning and implementing measures to prevent its introduction, thus providing a methodology for timely disease management and supporting the epidemiological surveillance and phytosanitary management programs.
Acknowledgments
We are grateful to DGSV-CNRF of SENASICA for directly supporting our research by letting us use equipment and reagents and obtain ATCC isolates.
REFERENCES
Altschul FS, Gish W, Miller W, Myers WE and Lipman JD. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403-410. https://doi.org/10.1016/S0022-2836(05)80360-2 [ Links ]
Ayyadevara S, Thaden JJ and Shmookler RJ. 2000. Discrimination of primer 3′-nucleotide mismatch by Taq DNA Polymerase during Polymerase Chain Reaction. Analytical Biochemistry 284: 11-18. https://doi.org/10.1006/abio.2000.4635 [ Links ]
Bonants JM Peter, Carroll CG, Weerdt M, Brouwershaven RI and Baayen PR. 2003. Development and validation of a fast PCR-based detection method for pathogenic isolates of the citrus black spot fungus, Guignardia citricarpa. European Journal of Plant Pathology 109: 503-513. https://doi.org/10.1023/A:1024219629669 [ Links ]
Cenis LJ. 1992. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Research 20 (9): 2380. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC312363/pdf/nar00083-0171.pdf [ Links ]
Crous PW, Verkleij GJM, Groenewald JZ, J Houbraken. 2005. Laboratory Manual Series No. 1: Fungal Biodiversity. CBS Fungal Biodiversity Centre. 425 pp. http://www.westerdijkinstitute.nl/News/Category/Laboratory%20Manual%20Series [ Links ]
Doyle JJ and Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11-15. https://webpages.uncc.edu/~jweller2/pages/BINF8350f2011/BINF8350_Readings/Doyle_plantDNAextractCTAB_1987.pdf [ Links ]
Everett RK and George Rees J. 2006. Species-specific PCR primers for Guignardia citricarpa and Guignardia mangiferae. New Zealand Plant Protection 59: 141-145. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.902.294&rep=rep1&type=pdf [ Links ]
Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95-98. http://brownlab.mbio.ncsu.edu/jwb/papers/1999hall1.pdf [ Links ]
Humber AR. 1997. Fungi: Preservation of cultures. Pp. 269-279. In: Lawrence A. Lacey (Ed.). Manual of techniques in insect pathology. Academic Press. 409p. https://doi.org/10.1016/B978-012432555-5/50015-4 [ Links ]
Hyun JW, Peres NA, Yi SY, Timmer LW, Kim KS, Kwon HM and Lim HC. 2007. Development of PCR assays for the identification of species and pathotypes of Elsinoë causing scab on citrus. Plant Disease 91: 865-870. https://doi.org/10.1094/PDIS-91-7-0865 [ Links ]
Luttrell SE. 1946. Black rot of muscadine grapes. Phytopathology 36: 905-924. https://naldc.nal.usda.gov/download/IND43894446/PDF [ Links ]
Manchester KL. 1995. Value of A260/A280 ratios for measurement of purity of nucleic acids. BioTechniques 19 (2): 208-210. https://jglobal.jst.go.jp/en/detail?JGLOBAL_ID=200902136544040775&rel=0 [ Links ]
Matveeva VO, Shabalina AS, Nemtsov AV, Tsodikov DA, Gesteland RF and Atkins FJ. 2003. Thermodynamic calculations and statistical correlations for oligo-probes design. Nucleic Acids Research 31 (14): 4211-4217. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC167637/ [ Links ]
Motohashi Keiichi, Shigeki Inaba, Kozue Anzai, Susumu Takamatsu and Chiharu Nakashima. 2009. Phylogenetic analyses of Japanese species of Phyllosticta sensu stricto. Mycoscience 50: 291-302. https://doi.org/10.1007/S10267-009-0487-Z [ Links ]
Okane I, Lumyong S, Nakagiri A and Ito T. 2003. Extensive host range of an endophytic fungus, Guignardia endophyllicola (anamorph: Phyllosticta capitalensis). Mycoscience 44 (5): 353-363. https://doi.org/10.1007/s10267-003-0128-x [ Links ]
Peres NA, Harakava R, Carroll CG, Adaskaveg EJ and Timmer WL. 2007. Comparison of molecular procedures for detection and identification of Guignardia citricarpa and G. mangiferae. Plant Disease 91 (5): 525-531. https://doi.org/10.1094/PDIS-91-5-0525 [ Links ]
Rinaldi AP, Paffetti D, Comparini C, Broggini ALG, Gessler C and Mugnai L. 2017. Genetic variability of Phyllosticta ampelicida, the agent of black rot disease of grapevine. Phytopathology 107: 1406-1416. https://doi.org/10.1094/PHYTO-11-16-0404-R [ Links ]
Rychlik W. 1995. Selection of Primers for Polymerase Chain Reaction. Molecular Biotechnology 3(2): 129-134. https://doi.org/10.1007/BF02789108 [ Links ]
Sambrook J and Russell DW. 2001. Molecular Cloning: A Laboratory Manual. 3ª ed. Ed Cold Spring Harbor Laboratory Press. Nueva York. USA. 2344. [ Links ]
Schoch LC, Seifert AK, Huhndorf S, Robert V, Spouge LJ, Levesque A and Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United State of America 109(16): 6241-6246. https://doi.org/10.1073/pnas.1117018109 [ Links ]
SENASICA. 2019. Pudrición negra de la vid (Guignardia bidwellii-Phyllosticta ampelicida). Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria. Dirección General de Sanidad Vegetal. Programa de Vigilancia Epidemiológica Fitosanitaria. México, D.F. Ficha Técnica No 63. 15p. Consultado en línea el 22/07/2019 a través del portal: https://prod.senasica.gob.mx/SIRVEF/ContenidoPublico/Fichas%20tecnicas/Pudrici%C3%B3n%20Negra%20de%20la%20Vid%20(Guignardia%20bidwellii).pdf [ Links ]
SIAP. 2018. Servicio de Información Agroalimentaria y Pesquera. Anuario estadístico de la producción agrícola: producción de vid. Consultado online en Abril de 2019 a través de https://nube.siap.gob.mx/cierreagricola/ [ Links ]
Stadhouders R, Paz DS, Anber J, Voermans J, Mes MHT and Schutten M. 2010. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. Journal of Molecular Diagnostics 12 (1): 109-117. https://doi.org/10.2353/jmoldx.2010.090035 [ Links ]
Ullrich IC, Kleespies GR, Enders M and Koch E. 2009. Biology of the black rot pathogen, Guignardia bidwellii, its development in susceptible leaves of grapevine Vitis vinifera. Journal für Kulturflanzen 61 (3): 82-90. [ Links ]
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M and Rozen SG. 2012. Primer3 - new capabilities and interfaces. Nucleic Acids Research 40(15): e115. https://doi.org/10.1093/nar/gks596 [ Links ]
Van der Aa H. 1973. Studies in Phyllosticta. Studies in Mycology 5: 1-110. [ Links ]
White JT, Bruns T, Lee S and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. PCR Protocols: a guide to methods and applications. 315-322. https://nature.berkeley.edu/brunslab/papers/white1990.pdf [ Links ]
Wicht B, Petrini O, Jermini M, Gessler C and Lodovico BGA. 2012. Molecular, proteomic and morphological characterization of the ascomycete Guignardia bidwellii, agent of grape black rot: a polyphasic approach to fungal identification. Mycologia 104(5): 1036-1045. https://www.ncbi.nlm.nih.gov/pubmed/22492405 [ Links ]
Wicht B, Jermini M, Gessler C and Lodovico BGA. 2014. Microsatellite markers for population studies of the ascomycete Phyllosticta ampelicida, the pathogen causing grape black rot. Phytopathologia Mediterranea 53(3): 470−479. http://www.jstor.org/stable/43871799 [ Links ]
Wikee S, Lombard L, Nakashima C, Motohashi K, Chukeatirote E, Cheewangkoon R, McKenzie EH, Hyde KD and Crous PW. 2013a. A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales). Studies in Mycology 76: 1-29. https://dx.doi.org/10.3114%2Fsim0019 [ Links ]
Wikee S, Lombard L, Crous WP, Nakashima C, Motohashi K, Chukeatirote E, Alias AS, McKenzie HCE and Hyde DK. 2013b. Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Diversity 60: 91-105. https://doi.org/10.1007/s13225-013-0235-8 [ Links ]
Ye Jian, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S and Madden LT. 2012. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. https://doi.org/10.1186/1471-2105-13-134 [ Links ]
Zhang Ke, Ning Zhang and Lei Cai. 2013. Typification and phylogenetic study of Phyllosticta ampelicida and P. vaccinii. Mycologia 105(4): 1030-1042. https://doi.org/10.3852/12-392 [ Links ]
Zuker M. 2003. mFold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406-3415. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC169194/ [ Links ]
Received: May 31, 2019; Accepted: July 28, 2019











 texto en
texto en 


