Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de fitopatología
versão On-line ISSN 2007-8080versão impressa ISSN 0185-3309
Rev. mex. fitopatol vol.37 no.2 Texcoco Mai. 2019 Epub 30-Set-2020
https://doi.org/10.18781/r.mex.fit.1902-6
Phytopathological notes
Identification of Tomato brown rugose fruit virus by RT-PCR from a coding region of replicase (RdRP)
1Dirección General de Sanidad Vegetal-Centro Nacional de Referencia Fitosanitaria. Carretera Federal México-Pachuca, Km 37.5, CP 55740, Tecámac, Estado de México, México
2Postgrado en Fitosanidad-Fitopatología. Colegio de Postgraduados Km. 36.5 Carretera México-Texcoco, CP 56230, Montecillo, Texcoco, Estado de México.
Currently, it is of strategic importance to have fast and accurate diagnostic tools that allow us to know if the material of interest is infected with a quarantine or economic important virus. For this reason in the present work we designed a pair of specific oligonucleotides for the detection of ToBRFV by RT-PCR, considering a region of the coding sequence of the RdRP located in the ORF1 and the methodology to detect it was standardized. Additionally, the amplicons were cloned and sequenced, the products were used to predict the evolutionary relationship between ToMMV, ToMV, TMV and ToBRFV by the Maximum Likelihood method. The results indicated that the oligonucleotides designed in the present work allows the identification of fast and specific ToBRFV.
Key words: Solanaceae; Tobamovirus; specific oligonucleotides; standardization
En la actualidad es de importancia estratégica contar con herramientas de diagnóstico rápido y preciso que permitan conocer si el material de interés está infectado con algún virus de importancia cuarentenaria o económica. Por tal motivo, en el presente trabajo se diseñó un par de oligonucleótidos específicos para la detección del ToBRFV por RT-PCR, considerando una región de la secuencia codificante de la RdRP localizada en el ORF1 y la metodología para detectarlo fue estandarizada. Adicionalmente, los amplicones fueron clonados y secuenciados, los productos fueron usados para predecir la relación evolutiva entre ToMMV, ToMV, TMV y ToBRFV por el método Maximum Likelihood. Los resultados indicaron que los oligonucleótidos diseñados en el presenté trabajo permiten la identificación de manera rápida y específica del ToBRFV.
Palabras clave: Solanaceae; Tobamovirus; oligonucleótidos específicos; estandarización
Tomato (Solanum lycopersicum) and chili crops (Capsicum annuum) are an economically important activity worldwide (Li et al., 2017). Historically, plants of the Solanaceae and Cucurbitaceae families have been affected by some species of the Tobamovirus genus, such as Tobacco mosaic virus (TMV) and Tomato mosaic virus (ToMV) (Dombrovsky and Smith, 2017). Tomato resistance to these viruses was introduced by introgression. However, the durability of resistance is compromised by the pathogen’s selection pressure (Dombrovsky and Smith, 2017; Maayan et al., 2018). Tobamoviruses are important because they are easily dispersed, mainly by mechanical means or through contaminated seed (Dombrovsky et al., 2017); the viral particles of tobamoviruses are extremely stable and remain infectious for several years (Dombrovsky and Smith, 2017). Recently, the presence of the Tomato brown rugose fruit virus (ToBRFV) was reported in Mexico, which affects tomato and chili crops (Cambrón-Crisantos et al., 2018). The infected plants produced fewer flowers and fruits, and developed necrosis on the peduncle and fruit calyx. In addition, yellow and necrotic areas, as well as rugosity, were observed on fruits (Salem et al., 2015).
To identify tobamoviruses, the use of oligonucleotides has been reported for universal detection, which were designed using an alignment matrix from regions located in the ORF1 that encodes the RdRP of 32 tobamoviruses (Li et al., 2018), but currently there are no reports on specific oligonucleotides. Therefore, the objective of this study was to develop a specific method for detecting ToBRFV, including the design of a pair of specific oligonucleotides to amplify an encoding region of the ToBRFV’s viral RdRP (ORF1) through reverse transcription polymerase chain reaction (RT-PCR) specifically considering the nucleotidic regions of the structural or catalytic motifs, as well as standardize the detection method.
To design the pair of specific oligonucleotides for ToBRFV the whole genome of the most representative species of the Tobamovirus genus reported by the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) was considered, including Tobacco mosaic virus (TMV, Virgaviridae) (NC_001367.1, FR878069.1, HE818443.1 and V01408.1), Tomato mosaic virus (ToMV, Virgaviridae) (NC_002692.1, KY967227.1, MF002490.1 and AF332868.1), Tomato mottle mosaic virus (ToMMV, Virgaviridae) (NC_022230.1, FX898034.1, KR824950.1 and KF477193.1); the genomes of two ToBRFV isolates (Virgaviridae) were also considered: the Tomato brown rugose fruit virus, an isolate from Israel TBRFV-IL (KX619418.1), and the Tomato brown rugose fruit virus, an isolate from Jordan (KT383474.1). A global alignment of the genomes was carried out using the BioEdit program version 7.0.5.3 (Hall, 1999) and the Clustal W algorithm in order to create a matrix and identify variable regions between them (Figure 1). The selected region was used to obtain the oligonucleotides sequences, which were then analyzed in silico using the OligoAnalyzer server version 3.1 (https://www.idtdna.com/calc/Analyzer/Home/Instructions) to predict their physical and chemical properties (Annex 1). The oligonucleotides were synthesized by Integrated DNA Technologies, Inc. (IDT).
To standardize the process, seven tomato plants were used as a ToBRFV positive control, and one of chili with putative ToBRFV symptoms collected in the state of Michoacan (Cambrón-Crisantos et al., 2018), which were stored at -70 °C in the facilities of the National Center for Phytosanitary Reference (CNRF, for its acronym in Spanish), Plant Health General Directorate, National Service for Plant Health, Safety and Agri-Food Quality (SENASICA, for its acronym in Spanish). The central midrib of leaves (0.5 cm wide approximately) was taken to prepare a composite sample that was then finely chopped. The total RNA extraction was performed using 100 mg of leaf midrib with the SV Total RNA Isolation System Start-Up® kit (PromegaTM), following the manufacturer’s specifications; RNA purity and concentration was quantified by spectrophotometry (Nano Drop 2000®, Thermo ScientificTM). We used as negative controls ten plants individually infected with the positive-sense single-stranded RNA genome virus: Rattail cactus necrosis-associated virus (RCNaV, Virgaviridae), Tobacco mosaic virus (TMV, Virgaviridae), Tomato mosaic virus (ToMV, Virgaviridae), Pepper mild mottle virus (PMMoV, Virgaviridae), Cucumber mosaic virus (CMV, Bromoviridae) and Papaya meleira virus (PMeV, Totiviridae), as well as with the DNA genome of Pepper golden mosaic virus (PepGMV, Geminiviridae) and Okra yellow mosaic Mexico virus (OYMMV, Geminiviridae).
The cDNA synthesis of the positive control was performed with random oligonucleotides (Random Hexamer, Invitrogen™), following the manufacturer’s instructions, under the following conditions: 1 cycle at 42 °C for 30 min, 1 cycle at 99 °C for 5 min, and finally at 12 °C for 5 min. For PCR, the specific oligonucleotides designed in the present study (ToBRFV-FMX and ToBRFV-RMX) were used (Table 1), which amplify a 475 pb fragment that encodes the RdRP region. The reaction mixture was prepared using 18.9 µL of molecular biology grade water (Invitrogen), 2.5 µL of 10X buffer (Invitrogen), 0.75 µL of MgCl2 (50 mM) (Invitrogen), 0.25 µL dNTP (10 mM) (Invitrogen), 0.75 µL of the ToBRFV-FMX primer (10 µM), 0.75 µL of the ToBRFV-RMX primer (10 µM), 0.1 µL of PlatinumTM Taq DNA polymerase (Invitrogen) and 1 µL of the cDNA in a final volume of 25 µL. The amplification program was as follows: 1 cycle at 95 °C for 5 min, 30 cycles at 95 ˚C for 30 s, at 55 °C for 30 s and at 72 ˚C for 40 s with a final extension at 72 ˚C for 7 min. Later, the PCR amplification of the viral RdRP fragment (positive plasmidic control) was cloned in the pGEM ®-T Easy cloning vector (Promega), and the insert ligation of 474 pb was performed in a total volume of 5 μL of reaction mixture; the components were: 2.5 μL of 2X ligation buffer, 0.5 μL of pGEM ®-T Easy vector, 0.5 μL of DNA Ligase T4 2X, 1 μL of the product to be cloned and 0.5 μL of water, keeping the mixture at 15 ˚C for 20 h. Subsequently, One Shot Mach1-T1 chemically-competent E. coli cells (Invitrogen) were transformed. For this, 150 µL of competent cells mixed with the 5 µL of the ligation reaction were used; the mixture was incubated in ice for 30 min and a thermal shock was applied at 42 ˚C for 2 min; then the tube was placed on ice for 2 min. 350 µL of LB medium were added and then the tube was incubated at 37 °C for 45 min. Finally, the mixture was sown on plates containing LB medium, 40 µL of ampicillin, 40 µL of IPTG and 40 µL of Xgal (20 mg mL-1) at 37 °C for 16 h. The pGEM/RdRP-ToBRFV (CP-1, ~3490 nt) construct obtained from this cloning was used as a positive control for standardizing the RT-PCR of the RdRP fragment. Later, the cloned fragments were sequenced in the Molecular Biology Laboratory of CNRF using the Applied Biosystems equipment model 3130, following the Sanger’s methodology.
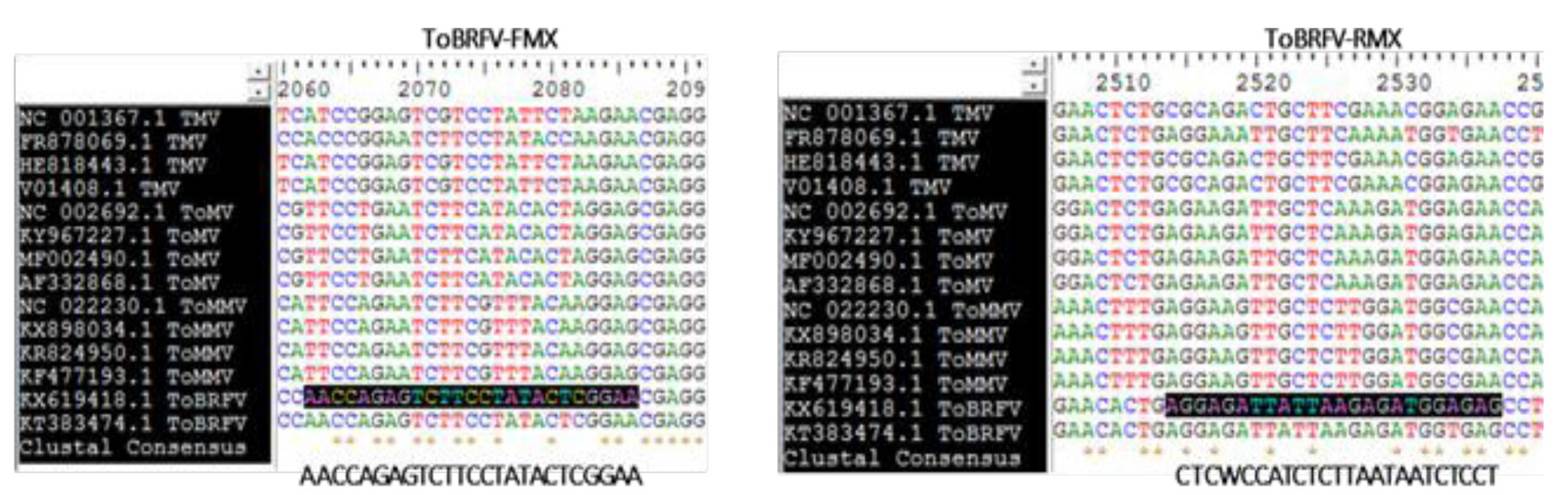
Figure 1 Selected region to design the specific oligonucleotides to detect ToBRFV. A) The Tobamovirus genomes TMV, ToMV, ToMMV and ToBRFV were considered.
Table 1 Characteristics of specific oligonucleotides to detect ToBRFV.
| Oligo (tamaño) | Secuencia 5´- 3´ | Tm | G/C | Amplicón |
|---|---|---|---|---|
| (°C) | (%) | |||
| ToBRFV-FMX (25 nt) | AACCAGAGTCTTCCTATACTCGGAA | 56.7 | 44 | 475 pb |
| ToBRFV-RMX (24 nt) | CTCWCCATCTCTTAATAATCTCCT | 51.5 | 37.5 |
The method standardization was achieved under the reaction conditions previously described. To determine the optimal annealing temperature, six annealing temperatures with a 4 °C difference between them (47, 51, 55, 59, 63 and 67 °C) were analyzed using six controls in a single concentration (100 ng µL-1): one positive plasmidic control to ToBRFV (CP-1), two biological controls infected with ToBRFV, To-P-100-2 and Ch-P-100-1 corresponding to tomato and chili, respectively; one matrix control per crop (CM-Ch-100-3 and CM-To-100-1) and one of reagents (NTC). The working range was calculated and determined using a positive plasmidic control to ToBRFV (CP-1) at a 1000 ng µL-1 concentration, and then serial dilutions were made in order to obtain three work concentrations (100, 10 and 1 ng µL-1) (Figure 2A). The lower limit of detection was established using the last dilution of the working range (1 ng µL-1), and then serial dilutions were made in order to obtain three work concentrations (0.1, 0.01 and 0.001 ng µL-1) (Figure 2B). The specificity was verified using RCNaV, TMV, ToMV, PMMoV, CMV and PMeV negative controls, as well as DNA genome such as PepGMV and OYMMV (Figure 2C).
Standardization data were used to detect ToBRFV from the plant samples collected in the state of Michoacan and stored at -70 °C; the cDNA synthesis was performed as previously described. The specific oligonucleotides designed in the present were used for PCR, which amplify a 475 pb fragment that encodes one RdPR region. The reaction mixture was prepared using 18.9 µL of molecular biology grade water (Invitrogen), 2.5 µL of 10X buffer (Invitrogen), 0.75 µL of MgCl2 (50 mM) (Invitrogen), 0.25 µL of dNTP (10 mM) (Invitrogen), 0.75 µL of the ToBRFV-FMX primer (10 µM), 0.75 µL of the ToBRFV-RMX primer (10 µM), 0.1 µL of PlatinumTM Taq DNA polymerase (Invitrogen) and 1 µL of the cDNA in a final volume of 25 µL. The amplification was performed at 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 40 s with a final extension at 72 °C for 7 min. The eight amplified sequences were visualized in 1.5% agarose gel (Invitrogen) and cloned in the pGEM ®-T Easy vector. The plasmids with the insert were sequenced in the Molecular Biology Laboratory of CNRF using the Sanger’s methodology and the Applied Biosystems equipment model 3130. The obtained sequences were assembled and edited to obtain 400 pb sizes and compared with sequences in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For this section, we used the eight samples used by Cambrón et al. (2018).
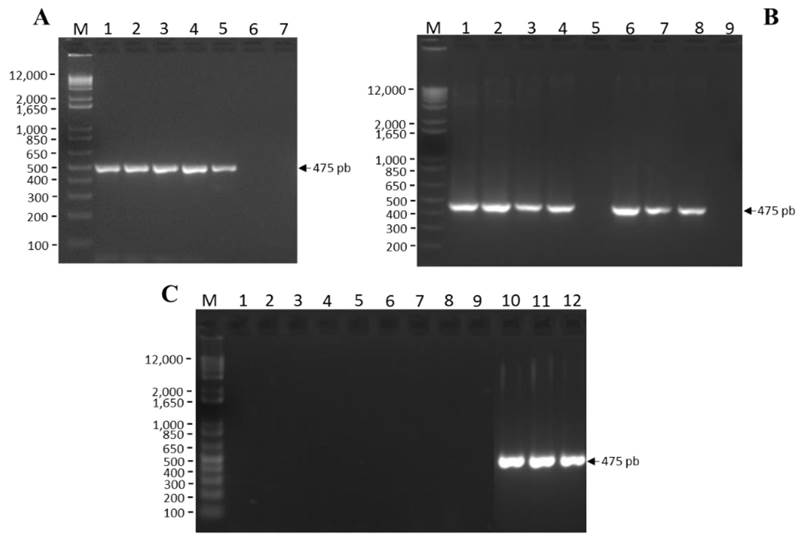
A) Annealing temperature: M= Molecular marker, 1= 47 °C, 2= 51 °C, 3= 55 °C, 4= 59 °C, 5= 63 °C, 6 = 77 °C and 7 = NTC.
B) Working range and lower detection limit: M= Molecular marker, 1= 1 000 ng μL-1, 2= 100 ng μL-1, 3= 10 ng μL-1, 4= 1 ng μL-1, 5= NTC, 6= 0.1 ng μL-1, 7= 0.01 ng μL-1, 8= 0.001 ng μL-1 y 9= NTC.
C) Specificity: M = Molecular marker, 1= RCNaV, 2= TMV, 3= CMV, 4= ToMV, 5= PMMoV, 6= PMeV, 7= PepGMV, 8= OYMMV, 9= NTC, 10= CP-1, 11= CM-Ch-100-3, 12= CM-To-100-1
Figure 2 Standardization of RT-PCR for the specific detection of ToBRFV.
Finally, to confirm the amplicon identity, an evolutionary analysis was conducted by sequence grouping in order to perform a local alignment using the MEGA program version 7.0.26 and the MUSCLE algorithm (Kumar et al., 2016). For the analysis, a matrix was created with sequences of Tobacco mosaic virus (TMV), Tomato mosaic virus (ToMV) and Tomato mottle mosaic virus (ToMMV). As external group to obtain the tree root, we used two sequences of Pepper mild mottle virus, an isolate from Huludao (PMMV) (MG515725.1) and Tobacco mild green mosaic virus, an isolate from Xiamen (ToMGMV) (JX534224.2). The Maximum Likelihood (ML) method based on the Tamura-Nei model with 1500 bootstrap replications was used. The initial trees for heuristic search were automatically obtained by applying the Neighbor-Join and BioNJ algorithms to a pairwise distance matrix using the Maximum Composite Likelihood (MCL) approach and selecting a topology with a higher log-likelihood value (Figure 3).
The in silico analysis of the ToBRFV-FMX and ToBRFV-RMX oligonucleotides showed to be specific to ToBRFV, based on the genome of several members of the Tobamovirus genus. In keeping with the predicted physical and chemical properties, the oligonucleotides are stable under PCR conditions. The amplicon of ~475 pb obtained by RT-PCR using the oligonucleotides designed in this study amplifies only ToBRFV in the positive control but not in samples infected with positive-sense single-stranded RNA (including several tobamoviruses) or DNA viruses, thus demonstrating that they are specific. The evolutionary analysis of the nucleotidic sequences corresponding to the RdRP fragment of the eight samples collected in Michoacan, Mexico, suggests an evolutionary relationship among TMV, ToMV and ToBRFV. Luria et al. (2017) mentioned an evolutionary relationship between TMV and ToBRFV. The results of the analysis conducted in this study using an ORF1 fragment that encodes the RdRP are in agreement with the results reported by Cambrón-Crisantos et al. (2018), where a 1052 pb PCR product was used with the F-3666 and R-4718 oligonucleotides, reported by Luria et al. (2017), which showed high nodal values in the tree they created. The standardization in this study showed that 55 ˚C is the optimal annealing temperature of the designed oligonucleotides, given that only the 475 pb amplicon was obtained. It was established that the working range for this methodology ranges from 1 ng µL-1 to 1000 ng µL-1, and that the lower limit of detection was ranges from 0.001 ng µL-1 to 0.1 ng µL-1. The oligonucleotides designed in this study can specifically identify the presence of ToBRFV in plant material, thus providing a methodology for timely detection, in less time and more specifically, compared to using general oligonucleotides for the Tobamovirus genus, without producing false positives with other single-stranded RNA genome (+ssRNA) viruses.
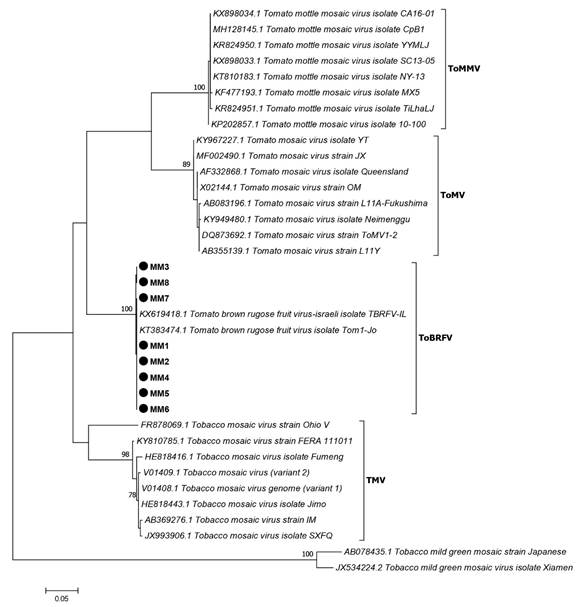
Figure 3 Evolutionary analysis of ToBRFV and viruses related to the Tobamovirus genus based on the alignment of the region corresponding to viral RdRP. The sequences included in the analysis are represented by the acronym of the virus and GenBank access numbers. The nodal values support the evolutionary relationship of the analyzed sequences.
LITERATURA CITADA
Cambrón-Crisantos JM, Rodríguez-Mendoza J, Valencia-Luna JB, Alcasio-Rangel S, García-Ávila CJ, López-Buenfil JA and Ochoa-Martínez DL. 2018. First report of Tomato brown rugose fruit virus (ToBRFV) in Michoacan, Mexico. Mexican Journal of Phytopathology, 37(1): 185-192. DOI: 10.18781/R.MEX.FIT.1810-5 [ Links ]
Dombrovsky A and Smith E. 2017. Seed transmission of Tobamoviruses: Aspects of global disease distribution. Advances in Seed Biology (pp. 234-260). DOI: 10.5772/intechopen.70244 [ Links ]
Dombrovsky A, Tran-nguyen LTT, and Jones RAC. 2017. Cucumber green mottle mosaic virus: Rapidly Increasing Global Distribution, Etiology, Epidemiology, and Management. Review in Advance of Phytopathol, 55(10): 1-26. DOI: 10.1146/annurev-phyto-080516-035349 [ Links ]
Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. 41: 95-98. [ Links ]
Li Y, Tan G, Lan P, Zhang A, Liu Y, Li R y Li F. 2018. Detection of Tobamoviruses by RT-PCR using a novel pair of degenerate primers. Journal of Virological Methods, 259: 122-128. DOI:10.1016/j.jviromet.2018.06.012 [ Links ]
Luria N, Smith E, Reingold V, Bekelman I, Lapidot M, Levin I, Elad N, Tam Y, Sela N, Abu-Ras A, Erza N, Haberman A, Yitzhak L, Lachman O and Dombrovsky A. 2017. A new israeli Tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE, 12(1): 1-19. DOI: 10.1371/journal.pone.0170429 [ Links ]
Maayan Y, Pandaranayaka EPJ, Srivastava DA, Lapidot M, Levin I, Dombrovsky A and Harel A. 2018. Using genomic analysis to identify tomato Tm-2 resistance-breaking mutations and their underlying evolutionary path in a new and emerging Tobamovirus. Archives of Virology, 163(7): 1863-1875. DOI: 10.1007/s00705-018-3819-5 [ Links ]
Salem N, Mansour A, Ciuffo M, Falk BW and Turina M. 2015. A new Tobamovirus infecting tomato crops in Jordan. Archives of Virology, 161(2): 503-506. DOI: 10.1007/s00705-015-2677-7 [ Links ]
Received: February 27, 2019; Accepted: April 23, 2019











 texto em
texto em 


