Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de fitopatología
versão On-line ISSN 2007-8080versão impressa ISSN 0185-3309
Rev. mex. fitopatol vol.37 no.2 Texcoco Mai. 2019 Epub 30-Set-2020
https://doi.org/10.18781/r.mex.fit.1901-3
Review articles
Molecular communication in the pathosystem Capsicum species -Phytophthora capsici
1 Laboratorio de Fisiología de la Interacción Planta-Patógeno, Especialidad de Fitopatología, Instituto de Fitosanidad, Colegio de Postgraduados, Km 35.5 Carretera México-Texcoco, Montecillo, Estado de México. C.P. 56230. Teléfono: 01 (595) 95-202-65 Extensión 1625. Fax: 01 (595) 95-202- 00 Extensión 1602. Correo electrónico: refzaid@hotmail.com
Phytophthora capsici is a phytopathogen that limits the production of vegetables worldwide. It is known to be the causal agent of the wilting of chili pepper, which affects the plantations of the genus Capsicum, causing almost complete losses. The pathosystem Capsicum spp. - P. capsici has been widely studied, but it is still far from being understood. Investigations on different chili pepper cultivars resistant to the oomycete suggest that most genotypes carry defense genes to confront the pathogen; however, the defensive capacity differs in intensity and speed. The specific resistance of some Capsicum species to P. capsici seems to be unrelated to R proteins, but rather mediated by a complex molecular dialogue. In some species of Capsicum, the growth regulators play an important part in this dialogue that leads the plant to express the genes related to defense, locally at first, by limiting the progress of the oomycete, and later, systemically, by preventing new points of infection. This revision carries out a critical analysis of the information available on the communication network established between chili plants and P. capsici, which defines the outcome of the interaction between the plant and P. capsici as resistant, tolerant or susceptible.
Key words: Molecular dialogue; plant immunity; plant-pathogen interaction; defense genes; growth regulators with the function of messenger molecules
Phytophthora capsici es un fitopatógeno que limita la producción de hortalizas en el mundo. Es reconocido como el agente causal de la marchitez del chile, que afecta a los cultivos del género Capsicum causando pérdidas casi totales. El patosistema Capsicum spp. - P. capsici ha sido ampliamente estudiado, pero se encuentra lejos de ser comprendido. Investigaciones sobre diferentes cultivares de chile resistentes al oomiceto, sugieren que la mayoría de los genotipos portan genes de defensa para afrontar al patógeno; sin embargo, la capacidad defensiva difiere en intensidad y velocidad. La Resistencia Especifica de algunas especies de Capsicum a P. capsici parece ser un fenómeno no relacionado a las proteínas R, sino mediado por un complejo diálogo molecular. En algunas especies de Capsicum los reguladores de crecimiento juegan un papel importante en este diálogo que lleva a la planta a expresar los genes relacionados con la defensa, primero de forma local, al limitar el avance del oomiceto, y después, de forma sistémica, al prevenir nuevos puntos de infección. En esta revisión, se hace un análisis crítico de la información disponible referente a la red de comunicación establecida entre las plantas de chile y P. capsici, misma que define el desenlace de la interacción entre la planta y P. capsici como resistente, tolerante o susceptible.
Palabras clave: Diálogo molecular; inmunidad vegetal; interacción planta-patógeno; genes de defensa; reguladores de crecimiento con función de moléculas mensajeras
In the process of evolution, pathogens have developed mechanisms to obtain nutrients from plants, causing disease or even the death of their hosts. In response, plants have developed physical and chemical barriers to impede the progress of the pathogen. These barriers may be pre-existing, inherent to the plant, such as waxy cuticles or alkaloids. Others are induced and activated by the interaction with phytopathogens, such as the reinforcement of cell walls with lignin or the production of toxic compounds, such as phytoalexins (Muthamilarasan and Prasad, 2013). These attack and defense mechanisms have evolved over time. Some pathogens, such as oomycetes, have even developed the ability to interrupt, interfere with, or evade the defense responses of plants (Brich et al., 2008; Lamour et al., 2012), which makes them devastating. On the other hand, plants have developed R proteins, which provide resistance to specific pathogens, and in their presence, lead to a fast reaction that interrupts the infection (Sanzón-Gómez and Zavaleta-Mejía, 2011; Gururani et al., 2012). The pathosystem formed by Capsicum spp.-Phytotphthora capsici is, in most cases, harmful to the host, which causes the wilting of the chili plant. Experimental evidence suggest the possible absence of R proteins that recognize the oomycete in the Capsicum genus (Smith et al. 1967; Reifschneider et al., 1992; Minamiyama et al., 2007); regardless, there is a strong resistance in certain genotypes (Mongkolporn and Tylor, 2011). The resistance of Capsicum to P. capsici has also been known to have a multigenic nature, and that the defense genes involved in the resistance to the oomycete are grouped in chromosome 5, although the intensity and the speed of the defensive response varies with the different varieties (Castro-Rocha et al., 2012; Barchenger et al., 2018). Likewise, the levels of resistance to P. capsici are modified by the presence of some biological control agents (Veloso and Díaz, 2012). The aim of this revision was to carry out a critical analysis of the state of the art of the pathosystem in question, which helped propose, in a synthesized manner, with the use of schemes, the complex communication networks that define the outcome of the interaction between the chili pepper and P. capsici as resistant, tolerant or susceptible.
The host solanaceae: Capsicum spp. The Capsicum genus is native to the tropics of the Americas. Its production and yield are limited by diverse phytopathogens such as bacteria (Xhanthomonas vesicatoria Doidge and Pectobacterium carotovorum Jones), fungi (Fusarium oxysporum Scltdl., Rhizoctocnia solani J.G. Kühn and Leveillula taurica (Lév.) E.S. Salmon), viruses (Pepper mottle virus, Pepper mild mottle virus, Cucumber mosaic virus and Tobacco mosaic Virus), nematodes (Meloidogyne spp. Goeldi and Nacobus spp. Thorne) and oomycetes (Pythium ultimum Trow and P. capsici) (Goldberg, 2001). Particularly, P. capsici has caught the attention of plant pathologists; named as “the destroyer of Capsicum plants,” the oomycete establishes a complex pathosystem by interacting with its host. However, some Capsicum cultivars have shown high levels of natural resistance, and in other cases, some varieties are able to resist the attack in the presence of biological control agent microorganisms (ACBs) with antagonistic or plant growth promotion abilities (Table 1). In both cases, the development or suppression of the disease is determined by a communication network, mediated by a variety of molecules.
Capsicum spp. molecules involved in the communication network. Just like other plants, Capsicum spp. displays three different types of molecules in the interaction with beneficial or pathogenic microorganisms. Those involved in the recognition of possible threats, those that carry the alert message and those expressed as defense responses.
Recognition molecules. Using Pattern Recognition Receptors (PRRs), plants detect and recognize physical, chemical and biological stimuli from the surroundings (Muthamilarasan and Prasad, 2013). The Receptor-Like proteins with leucine rich repeats domains and a C-terminal bound to the membrane (RLPs); and RLKs, which possess an N-terminal extracellular domain, a transmembrane domain and a C-terminal kinase domain inside the cell are the most widely described receptors. Both receptors are found in the membrane, perceive foreign molecules, and send an alert to the inside of the cell. Thus, PRRs detect Microorganism (MAMPs) and Pathogen (PAMPs) Associated Molecular Patterns such as flagellin domains, exopolysaccharides, products of the Type 3 Secretion System (T3SS) and its peptidic domains, peptidoglycans and molecules related to the quorum sensing such as the N-acyl-homoserin-lactones (AHL) (negative Gram bacteria) or oligopeptides (positive Gram bacteria). These molecular patterns reveal the presence of bacteria in the plant. The presence of fungi is revealed by molecules such as chitin, β-glucans, lypoproteins, specific glycosylated proteins, ergosterol and olygomannosides. In oomycetes such as P. capsici, the domains of PEP-13 of the transglutaminase, PcNPP1 type proteins, elicitins as capsicein and lipids as arachidonic acid, warns its presence to the host plant (Bent and Makey, 2007; Muthamilarasan and Prasad, 2013; Vidhyasekaran, 2014). In 2010, Yi and coworkes described a RLK-type receptor in Capsicum, related to the delay of the Hypersensitive Response (HR). Despite the ability of the PRRs to detect the presence of invasive microorganisms, some phytopathogens have developed different strategies to establish themselves and colonize plant tissues. The pathogens that significantly affect agricultural production are able to block, interfere or evade plant defense responses (Lamour et al., 2012). On the other hand, plants have developed specialized molecular receptors such as R proteins (product of resistance genes), capable of triggering a violent defense response. R proteins remain in the cytosol in order to directly recognize the effectors (receptor-ligand model), or it may be bonded to the target molecule to recognize the action of the effector on it (guardian model) (Brich et al., 2008), and therefore activate the resistance mediated by the RH. The R proteins consist of a domain with leucine-rich repeats (LRR) and a nucleotide biding domain (NB) coupled to a 1-Toll-Interleucin receptor (TIR-NB-LRR) (Gururani et al., 2012).
The R genes of Capsicum are well-known: gene Bs2 confers resistance to Xanthomonas campestris (recognizes AvrB2), CaMi recognizes Meloidogyne incognita (it recognizes different molecules related to the plant-nematode interaction), pvr1 and pvr2 recognize the Potato virus Y (they recognize Vpg), and L1, L2 and L3 recognize the Turnip mosaic virus (they recognize a protein of the capsid) (Gururani et al., 2012). Cannon et al. (2002) suggest the possible presence of more R genes distributed in the species of Capsicum. Various authors have concentrated on the search for specific R genes involved in the resistance to P. capsici in resistant lines, such as the serrano Criollo de Morelos 334 (CM334), ACC 2258 and Smith 5 genotypes (Smith et al. 1967; Reifschneider et al., 1992; Minamiyama et al., 2007). Results are contradictory and lines with a stable resistance have not been obtained (Mongkolporn y Tylor, 2011). Based on this information, it is possible to speculate that the phenomenon of resistance is not related to the presence of R proteins. Other authors agree in attributing the resistance to a multigenic origin, in which a prompt, intense and coordinated response leads to a local RH, along with the overexpression of genes such as PAL, EAS and PR genes such as POX, GLU and CHI, mainly (Fernández-Herrera et al., 2012; Sudisha et al., 2012; Villar-Luna et al., 2015). The experimental evidences suggest that, in general, Capsicum plants are able to recognize the oomycete P. capsici, and activate the same defense responses, althought only some cultivars display resistance (Castro-Rocha et al., 2012).
Table 1 Diversity of biological control agents used to protect the plant against the wilting of chili and its action mechanisms implied and the resulting effect of the interaction.
| Agente de Control Biológico | Variedad de Chile | Mecanismo Reportado en: | Efecto | Referencia | |
|---|---|---|---|---|---|
| Antagonista | Planta | ||||
| Bacillus amyloliquefaciens, B. thuringensis | Hibrido SV3198HJ | Antibiosis | - | Inhibición en germinación de zoosporas | Ley-López et al.,2018. |
| Penicillumsp. | Anaheim | Micoparasitismo y competencia | - | Desintegración de hifas | Jiménez-Camargo et al.,2018 |
| Trichoderma hamatumy Pseudomonas aeruginosa | Landung | Micoparasitismo y competencia | - | Reducción de la incidencia de la enfermedad | Chemeltorit et al.,2017. |
| Paenibacillus polymixa | Supermanitta | Antibiosis e ISR | POX, PPO, PAL, SOD | Promoción de la salud vegetal | Xu y Kim 2016. |
| Streptomyces plicatus | California wonder | Antibiosis, hiperparasitismo | - | Reducción de la incidencia de la enfermedad | Chen et al.,2016 |
| Fusarium oxysporum | - | ISR y competencia | PR1, CHI, SC | Promoción de salud vegetal | Veloso y Díaz, 2012 |
| Trichoderma ovalisporum, T. hamatum, T. theobromicola, T. stilbohypoxyl, T. carbbaeum | Bugang | Micoparasitismo y antibiosis | CHI, PR4, EAS, SC | Reducción en el desarrollo de la enfermedad | Bae et al.,2011 |
| Xylaria poitei | Mirasol | Competencia | - | Protección de la planta | Ramos et al.,2010 |
| Glomus mosseae, G. etunicatum, G. fasciculatum, G. margarita | Charliston Bagci | ISR y competencia | PAL, CHI, GLU, capsidiol | Mayor desarrollo de raíces y resistencia a la enfermedad | Ozgonen et al.,2009 |
CHI Quitinasas; EAS Epi-5-Aristoloqueno sintasa; GLU Glucanasas; PAL Fenilalanina amonio liasa; POX Peroxidasas; PPO Polifenoloxidasas; PR1 y PR4 Proteinas relacionadas a la patogénesis; SC Sesquiterpeno cyclasa; SOD Superóxido dismutasa / CHI Chitinases; EAS Epi-5-Aristoloquene synthase; GLU Glucanases; PAL Phenylalanine amonnia lyase; POX Peroxidases; PPO Polyphenoloxidases; PR1 and PR4 Proteins related to pathogenesis; SC Sesquiterpene cyclase; SOD Superoxide dismutase
Signal Molecules. If the plant recognizes a potential risk, it transmits and amplifies the signal through a series of diverse molecules. These may be anywhere from reactive species of oxygen (ROs), proteins (Ca+ dependant proteins , mytogen-activated kinase proteins), growth regulators that function as signal molecules (jasmonic acid, ethylene, calicylic acid), up to complex receptors and even transcription factors (WRKY, CaRFLP, MYC) (Yi, et al., 2010; Vidhyasekaran, 2014).
Response Molecules. Resistance in Capsicum species is far from being fully understood; in some cultivars it depends on the density of the inoculum or the plant’s physiology. However, several researchers have demonstrated the important role of some products of the genes related to defense in chili plants. Among these are the family of genes that codify for phenylalanine ammonia lyase (PAL), involved in the synthesis of phenoles, which are precursors in the process of cell wall reinforcement and defense by toxicity, and is also a key enzyme for the synthesis of salicylic acid (Vidhyasekaran, 2014). Genes such as EAS, involved in the synthesis of the phytoalexin capsidiol and POX, involved in the processes of lignification, contribute to establishing chemical and physical barriers, respectively (Villar-Luna et al., 2015; Villar-Luna et al., 2017). In addition, the genes of proteins related to pathogenesis (PRs) such as PR2 (glucanases) and PR3 (chitinases), degrade the cell walls of the pathogens (Dahiya et al., 2006; Hardham and Shan, 2009). The quick accumulation of these molecules in the plant makes it a hostile medium that limits the biological cycle of the pathogen. Although these are the main genes involved in defense, there are many others that participate in the response of the plant and that are regulated positively or negatively. For example, the family of genes that codify for the enzyme hydroxymethylglutaryl-coenzyme A reductase (HMGR), involved in changing the path of the mevolanate towards the synthesis of isoprenoid compounds, such as steroles (HMG1) that favor the development of the oomycete, or the production of sesquiterpenic phytoalexins, such as (HMG2 and HMG3) that limit its development (Villar-Luna et al., 2017).
The pathogen: Phytophthora capsici. The range of P. capsici hosts includes species of Capsicum (C. annuum, C. baccatum, C. chinense, C. frutesens and C. pubescens), cucurbits, eggplants and tomatoes, as well as green beans and broad beans (Lamour et al., 2012). The parasitic success of P. capsici is due to several evolutionary advantages: 1) its mobile, flagellated zoospores equipped with receptors increase their ability to disseminate and search (Bishop et al., 2002); 2) its resistance structures (thick-walled oospores) can survive for up to 4 years in the soil, and are the main source of primary inoculant (French et al., 2007); 3) its ability to break up the cell wall by secreting polygalacturonases, pectin methylesterases and pectate lyases (Feng et al., 2010), as well as the cell membrane by secreting capsicein (Nespoulous et al, 1999); and 4) its hemibiotrophic habits, the ability to feed off living and dead tissue (Hardham and Shan, 2009).
Molecules of P. capsici involved in the interaction with Capsicum. P. capsici can activate or suppress the basal immunity of plants by producing specialized molecules that may promote a favorable environment for its development and reproduction (Hardham and Shan, 2009); these molecules are known as PAMPs (Pathogens Associated Molecular Patterns) and effectors (Kamoun, 2006; Bent and Mackey, 2007).
PAMPs. PAMPs and MAMPs are preserved molecules that contribute to the biological fitness of microorganisms (Thomma et al., 2011). They are also known as elicitors, and are recognized by the receptors of the plant cell by activating the immunity of the plant; this event is known as Pathogen-Triggered Immunity or Microorganism-Triggered Immunity (PTI o MTI) (Torto et al., 2009). PAMPS of P. capsici that interact with Capsicum are: Peptide PEP-13, a glycoprotein that activates the synthesis of phytoalexins and of ROs (Tör, 2008). PcPNPP1, that acts as a virulence factor, helps it change its habits from biotrophic to necrotrophic, and induces a programmed cell death (PCD) (Jupe et al., 2013). Capsicein, a compound that encapsulates the ergosterol of the host cell membrane and translocates it to the cells of the pathogen, and also triggers diverse responses (Nespoulous et al, 1999). Arachidonic Acid, that induces an increase in the concentration of ethylene and elicits RH (Bostock et al., 2011).
Effectors. The effect of these molecules depends on the genotype of the plant. In susceptible plants, the effector acts as a factor of virulence, modifying the plant cell structure and function inducing the disease. In resistant plants (with specific R genes), the same effector can activate the immunity of the plant (ETI) acting as a factor of avirulence (Win et al., 2012). In addition, the non-pathogenic symbionts produce effectors to establish parasitic relationships, whether mutualistic or commensalistic (Torto et al., 2009). The effectors produced by P. capsici are RXLR, proteins with a highly preserved N-terminal domain and an RXLR motive, in which X participates in its translocation to the host cell; this effector modulates and suppresses the defense responses during the biotrophic phases, and acts as an enhancer during pathogenesis. At least 400 putative genes related to these molecules have been identified in P. capsici (Brich et al., 2008; Lamour et al., 2012). Crinklers contain an N-terminal domain, a highly conserved LXLFLAK motive and a C-terminal domain (promoter of virulence); they act on proteins in the nucleus possibly involved in the transportation of nucleic acids. These molecules are associated to necrosis and epinasty; 80 genes have been identified that seem to codify for crinkler-type effectors in P. capsici (Stam et al., 2013). Other effectors, such as the EPIs similar to the glucanase inhibitors described in P. sojae and cystatine-type proteases described in P. infestans (Tian et al., 2016), may also be produced by P. capsici, since the presence of EPI genes has also been found in their genome.
Immunity activated by MAMPs or PAMPs and the induction of systemic resistance in Capsicum spp. The interaction between Capsicum spp. and P. capsici takes place through a dialogue mediated by the molecules mentioned above. The result of this dialogue depends widely on the biological fitness of the organisms (ability to respond to stimuli), environmental factors, and even on multiple specific factors. For example, the agronomic manage, the phenological stage in which the interaction takes place, or even the type of microbial populations present in the pathosystem at that moment. The sum of the particularities under which a pathosystem takes place is directly related with the different outcomes in which the interaction can end.
The first possible outcome is that the immunity is activated by MAMPs or by PAMPs (MTI or PTI). The plant behaves as resistant against plant pathogens, endophytes, and non-adapted or attenuated pathogens, or as tolerant resulting in asymptomatic infections. In this process, MAMPs or PAMPs stimulate receptors RLPs and RLKs (including CaRLK1). These activate the G proteins of the membrane that act as a switch that “turns on” the ion pumps (Vidhyasekaran, 2014). Thus begins an ionic flow through the membrane introducing H+ and Ca2+ and pumping Cl- and K+ to the outside (Sanzón-Gómez and Zavaleta-Mejía, 2011). The increase of Ca2+ ions in the cytosol activates signaling processes along with NADPH oxidase, increasing the production of ROs (H2O2, O2 - and OH-), and preparing the cell for a possible PCD (Glowacki et al., 2011). In addition, Ca2+ activates proteins such as calmodulins (CaMs), type B calcineurins (CLBs) and calcium-dependant kinases (CDPKs) (Vidhyasekaran, 2014). These proteins phosphorilate the MAP-Ks, carrying the signal downstream to the type WRKY transcription factors that recognize the W boxes (TTGAC[C/T]) of the genes related to defense; then, a reprogramming in the transcription leads to cellular metabolic changes in the infection zone(Jingyuan et al., 2011) (Figure 1).
The Capsicum cells around the area of infection accumulate toxic substances such as phenolic compounds (synthesized in the route of the phenylpropanoids, where the enzyme PAL is key) or products of its oxidation, and phytoalexins such as capsidiol (synthesized by EAS).
There is also an increase in the synthesis of PRs involved in the dissociation of the cell walls of the pathogens (GLU and CHI), and in the reinforcement of cell walls of the host by lignification (POX) (Castro-Rocha et al., 2012). Along with the defense response, the stimulus on the receptor CaRLK1, specific to Capsicum, increases the expression of enzymes superoxide dismutase (SOD), apparently to alleviate the oxidative stress and to delay PCD, allowing other processes to take place. The radical superoxide (H2O2) and nitric oxide (NO) are not essential to activate RH, but when a necrotrophic pathogen, neither adapted nor weakened, endophytes or ACBs stimulate the CaRLK1 receptors, these increase the production of SOD, disgregating the H2O2 and increasing the concentration of the free oxygen radical (O2 -). Reacting O2 - with the NO forms peroxynitrite (ONOO-), that prevents the interaction between NO and H2O2 and delays PCD (Figure 1). In addition, ONOO is toxic for some microorganisms, including necrotrophic pathogens (Yi et al., 2010).
Basal defense (MTI/PTI) is effective against some necrotrophic pathogens or invasive microorganisms not related to the plant, although it only covers the area around the infection. Simultaneously to MTI/PTI, signaling events mediated by the plant growth regulators as jasmonic acid (JA) and ethylene (ET) take places to induce systemic resistance (ISR) in Capsicum plant. Naturally, phospholipase A (PLA) releases α-linoleic acid (α-lin) from the membrane, although the damage caused by the entrance of pathogens or ACB causes a greater release of this acid. The presence of α-lin and the uprising concentration of ROs timulate the octadecanoid pathway, in which lypoxygenase (LOX) and other enzymes turn free α-lin into oxylipins (Bertoni, 2012). Out of the oxylipins produced, the most important group is that of JA, which sends the signal to the distant cells, in which a complex receptor (JAR) formed by three molecules (F-box protein COI1, zinc finger protein JAZ and inositol pentakisphosphate) receives the alert. It is still unclear if the signaling process continues downstream via MAP-Ks; however, the signal reaches the nucleus activating a family of transcription factors formed by a basic protein of the type helix-loop-helix (MYC factors), which in turn, activate the expression of genes related to the defense (Vidhyasekaran, 2014). Some of the genes expressed in solanaceae by JA signal correspond to those that encode for PRs as PR 1, PR 5 (thaumatin like proteins) and PR 6 (proteinase inhibitors) (Sudisha et al., 2012) (Figure 2A). At the same time, the ET signaling path takes place to complete the activation of the ISR. The increase in the concentration of ROs activates the transcription of two key enzymes for the synthesis of ET: ACC synthase (ACS) and ACC oxidase (ACO); in addition, CDPKs activated by the Ca+ increase the activity of these enzymes, triggering the synthesis of ET.

Figure 1 MAMPs or PAMPs triggered immunity in Capsicum plants (Yi et al., 2010; Glowacki et al., 2011; Jingyuan et al., 2011; Sanzón-Gómez y Zavaleta-Mejía, 2011; Castro-Rocha et al., 2012; Vidhyasekaran, 2014). ApSOD Super oxide dismutase in the apoplast; CaMs Calmodulines; CaRLK Receptor-like kinases in Capsicum related to HR delay; CBLs B type Calcineurines; CDPKs Calcium Dependent Kinases; CHI PR3-Chitinases; Cpsdl Capsidiol Synthesis; CySOD Superoxide dismutase in the cytosol; EAS 5-epi-aristolochene syntase; GLU PR2-Glucanases; GP G Proteins; IONP Ion Pumps; HRD HR Delayed; LDR Local Defense Responses; Lig Cell-wall lignification process; MAP-Ks Mitogen Activated Protein Kinases; MTI MAMPs Triggered Immunity; NADPH Nicotinamide adenine dinucleotide phosphate;NADP reduced NADPH; NADPHOx Enzyme NADPH oxidase; NO Nitric oxide; ONOO- Peroxinitrite; PAL Phenilalanine Ammonia Lyase; Phn Phenoles synthesis; POX PR9-Peroxidases; PRRs Pattern Recognition Receptors; PTI PAMPs Triggered Immunity; RLK Receptor-Like kinase; RLP Receptor- Like protein; ROs Oxygen Reactive Species; WRKY WRKY type transcription factors.
The growing concentration of ET carries the alert to distant cells, where it is recognized by trans-membrane receptors (ETR) composed of five molecules (ETR1, ETR2, ERS1, ERS2 and EIN4). The ETR molecules interact amongst each other and with CTR1 (with Raf kinase considered as a MAP-K) to activate downstream the signaling via MAP-Ks towards the nucleus (Vidhyasekaran, 2014). Later, the transcription factor CaERLFP receives the signal and recognizes the GCC boxes of the genes of the PRs (Lee et al., 2004), inducing the expression of PRs as PR 2 (GLU), PR 3 (CHI), PR 7 (endoproteinases), PR 9 (POX) and PR 12 (defensins) (Sudisha et al., 2012) (Figure 2A). The activation of the ISR varies in intensity according to the circumstances of the interaction, and does not always result in the systemic activation of the defense responses, but it may result in a higher ability to respond faster and more intensely against the attack by phytopathogens in the surrounding tissues (priming effect) (Conrath, 2009). ISR is effective against a wide range of phytopathogens, yet it is more complex than explained here, since the signaling paths that participate in their activation also regulate in a positive and negative manner a considerable amount of genes that may be involved in the process of induction of resistance.

Figure 2 Induction of systemic resistance, effector triggered immunity and systemic aquired resistance in Capsicum plants. (A) Induction of systemic resistance (ISR); (B) effector triggered immunity (ETI); (C) systemic acquired resistance (SAR) (Lee et al., 2004; Lee y Hawng, 2005; Conrath, 2009; Yi etal., 2010; Sanzón-Gómez y Zavaleta-Mejía, 2011; Bertoni, 2012; Gururani et al., 2012; Sudisha et al., 2012; Veloso et al., 2014; Vidhyasekaran, 2014). αLin α-Linoleic acid; Act. Activates to; ACS ACC Syntase; ACO ACC Oxidase; AvrBS2 R Protein product of resistance genes; BS2 Avirulence protein (Effector); CaERLFP Transcription factors activated by ET signal; CaMs Calmodulines; CDPKs Calcium dependent kinases; CHI PR3-Chitinases; CTR1 Raf kinase considered as MAP-K; CySOD Superoxide dismustase in the cytosol; Cyt Cytoplasm; ET Ethylene; ETI Effector Triggered Immunity; ETR Ethylene receptors; GLU PR2-Glucanases; GP G Proteins G; HR Hypersensitive Response; HRTNC HR triggered in neighbor cells; ICS Isocorismato Sintasa; IONP Ion Pumps; ISR Induce Systemic Resistance; JAR Jasmonic acid receptors; JA Jasmonic acid; LDR Local Defense Responses; LOX Lipoxigenase; RLK Receptor-Like Kinases; RLP Receptor-Like protein; MAPKs Mitogen activated protein kinases; MYC Transcription factors activated by JA signal; NADPHOx NADPH oxidase enzyme; NO Nitric oxide; NPR1, 3, 4 High affinity to SA receptors; Nuc Nucleus; OctPth Octadecanoid pathway; PAL Phenilalanine ammonia lyase; PLA Phospholipase A; Phn Phenol synthesis; PR1 Protein related to pathogenesis from Group 1; PR5 Thaumatin; PR6 Proteases inhibitor; PR7 Endoproteinases; PR12 Defensins; POX PR9-Peroxidases; ROs Reactive Oxygen species; SA Salicylic Acid (if + appear as suffix accompanying a sign denotes the increase in the concentration of this compound, a greater number of + denotes a greater increase); SAR Systemic Acquired Resistance; SDR Systemic Defense Responses; Ub Marked by ubiquitination; VROB Violent ROs Burst; TGA Transcription factors activated by NPR signal; * Goes to, ? Signaling pathway under investigation.
A second possible outcome is the development of the disease. Factors such as a nutritional deficiencies, the absence of beneficial microorganisms, the phenological state, the inoculum potential and management practices, all directly affect the expression of MTI/PTI, ISR and even the ACBs. The combination of non-optimum conditions for the plant’s development and the presence of an easily adaptable pathogenic genotype, leads to the development of the disease. In this case, P. capsici enters the tissue directly via the formation of an appressorium, but in both cases, by the secretion of enzymes (Feng et al., 2010; Castro-Rocha et al., 2012). During the biotrophic phase, the hyphae remain in the apoplast and the uptake of nutrients and secretion of virulence factors is carried out with a specialized haustorium invaginated into the cell membrane of the host. In this point, P. capsici secretes molecules such as capsicein to obtain the sterols contained in the host cell membrane (Nespoulous et al, 1999). However, the presence of this and other molecules such as Pep-13 and arachidonic acid (PAMPs), alert the plant and activate PTI. In response, P. capsici releases the effectors RLXLR and Crinklers (CRN) to evade the defense responses. Diverse contributions reveal that in incompatible interactions, these effectors accumulate in the nuclei of the host, blocking the expression of defense genes (Stam et al., 2013). In this way the modulation of the expression of the local genes suppresses the PTI during the biotrophic phase (Kamoun, 2006). This event helps the oomycete feed itself and mature. Later, it produces the protein PcPNPP1, which first changes the behaviour of the phytopathogen from biotrophic to necrotrophic; and finally, when secreted to the host’s tissue, it induces PCD and necrosis, allowing the pathogen to colonize the tissue and cause the disease (Lamour et al., 2012; Jupe et al., 2013).
Effector-triggered immunity (ETI) and systemic resistance in Capsicum spp. In order to hypothetically explain the specific resistance to P. capsici, it is necessary to be based on the knowledge on the way R proteins work when faced with biotrophic pathogens such as Xanthomonas campestris or Meloidogyne incognita in resistant Capsicum cultivars. The R proteins reported for Capsicum correspond to the recognition of bacteria, nematodes and viruses with biotrophic habits; for the moment, no R proteins have been reported for fungal phytopathogens or oomycetes with necrotrophic or hemibiotrophic habits. When the R proteins of Capsicum recognize an effector, they generate a dramatic increase in the concentration of ROs via the signaling of Ca+, which inhibits the local expression of enzymes such as SOD, and it favors a balance between H2O2 / NO, activating the RH (Yi etal., 2010; Gururani et al., 2012). Ca+ also activates the defense responses via MAP-K and the expression of the isochorismate synthase (ISC) via CaMs. The expression of PAL results in the accumulation of phenoles and phytotoxic compounds that accompany the RH and the expression of ISC in the synthesis of SA (Lee and Hawng, 2005; Sanzón-Gómez and Zavaleta-Mejía, 2011). This event characteristically culminates in a fast and intense PCD, and is known as effector-triggered immunity (ETI) (Figure 2B). The growing production of SA leads to its translocation to distant cells, where it is received by three receptors: NPR1 (medium affinity to SA), NPR3 (low affinity) and NPR4 (high affinity). Upon the arrival of the SA to the distant cell, NPR1 migrates from the cytosol to the nucleus, where NPR4 joins it and marks it by ubiquitination for its degradation, although, if the concentration of SA increases, NPR4 joins SA and releases NPR1. The continuous increase on the concentration of SA leads to the union between NPR1 and a molecule of this growth regulator; this complex acts as a cofactor of the TGA transcription factor, activating the defense responses related to the systemic acquired resistance (SAR). Finally, at high concentrations of SA, NPR3 joins the complex SA-NPR1, which activates the PCD in neighboring cells (Veloso et al., 2014) (Figure 2C). The activation of ETI and SAR in Capsicum involves the local and systemic activation of certain defense responses while RH takes place, the most important of which are the deposition of callose, the lignification of cell walls and the increase in the concentration of PRs such as PR-1 (defensin), PR-2 (glucanases), PR-3 (chitinases), PR-5 (osmotine), PR-9 (peroxidases), PR-10 and PR-13 (thionines) (Lee and Hwang, 2005). ETI is effective against phytopathogens that have Avr genes, corresponding to the R proteins in Capsicum, and the defense of the SAR is long-lasting and effective against a wide range of phytopathogens.
Specific resistance of Capsicum spp to Phytophthora capsici. In order to understand the third outcome (the specific resistance of Capsicum to P. capsici) it is necessary to consider the differences between resistant and susceptible cultivars. 1) In susceptible cultivars, the expression of enzymes that degrade ROs such as POX, SOD and CAT is accelerated and contributes to the delay of the PCD; on the other hand, in resistant cultivars, the expression of these enzymes is slow and they are suppressed locally by the violent increase in the concentration of ROs (Ueeda et al., 2006; Yi et al., 2010). 2). In resistant cultivars, the octadecanoic path is always active and it generates drastic increases in the levels of JA (Ueeda et al., 2006), unlike the susceptible cultivars. 3) There is a correlation between the concentration of capsidiol, the total of the area with necrosis and the invasion of the oomycete; in resistant cultivars, large amounts of capsidiol concentrate in small necrotic areas, inhibiting the progress of P. capsici; in susceptible plants, the amounts of this phytoalexin are lower, necrotic areas are large, and there is more infection in the tissue (Egea et al., 1996; Villar-Luna et al., 2015). 4) Resistant cultivars display a greater expression of genes PAL, HMG2, HMG3 and EAS associated to a greater activity of the corresponding enzymes; this is reflected in a higher concentration of phenols, toxic for the oomycete and precursors of lignin and synthesis of phytoalexins (López-Martínez et al., 2011; Villar-Luna et al., 2015; Villar-Luna et al., 2017) and SA (Lee and Hawng, 2005).
Based on the above, we propose that the Specific Resistance of Capsicum to P. capsici (RECP) begins with the recognition of PAMPs by receptors RLPs and RLKs, just like PTI, except that in this case, there is a rapid increase in the concentration of ROs that locally inactivates the enzymes that dissociate H2O2. The accumulated H2O2 reacts with the NO activating the RH (between 12 h and 24 h) (Villar-Luna et al., 2009). During this time, the local defense responses become activated, as in the PTI. In resistant cultivars, the octadecanoic and ICS paths generate drastic increases of the growth regulators involved in the signaling (JA-ET o SA). Unlike ISR and SAR, in which signaling paths seem to be antagonistic, in RECP there is a coordinated process in which each growth regulator plays an important part in a given time (Ueeda et al., 2006). Initially, JA reaches a maximum concentration approximately 2 h after recognition, making the surrounding tissue sensitive; the ET reaches the maximum level after 6 to 12 h, inducing the accumulation of PRs in neighboring cells (Kim and Hwang, 2000). Overall, these growth regulators induce the cell wall reinforcement process carried out by POX, and the systematic expression of GLU and CHI prevents new points of infection (Jung et al., 2005), as in ISR. Once the defense responses become activated, JA levels decrease after 6 h, and in ET, after 12 h. On the other hand, the concentration of SA increases after 6 h, and reaches its highest point after 12 and 24h, coinciding with the activation or HR (Ueeda et al., 2006, Villar-Luna et al., 2009). In this stage of the response, the cultivars that behave as resistant show necrotic areas in the points of infection with a high concentration of capsidiol and phenols (López-Martínez et al., 2011; Ozgonen et al., 2009; Villar-Luna et al., 2009), the surrounding cell walls are thickened and protected by the concentration of PRs (Jung et al., 2005). Afterwards, the increase in SA stimulates the systemic expression of the defense genes associated to the SAR; furthermore, if the level of SA reaches high concentrations, it activates the PCD in cells surrounding the infected area, making the progress of the phytopathogen more difficult (Veloso et al., 2014) (Figure 3).
The process of the RECP described was based on the experimental evidence that deal with the interaction between resistant, tolerant and susceptible cultivars with the oomycete and with ACB, in order to propose a basis for the promotion of investigations that tackle the distinctive features of this pathosystem. Despite the experimental evidence being extensive and despite them helping relate the information to explain, up to a certain point, the resistance of Capsicum to P. capsici, there are still many questions, such as, 1) What makes resistant cultivars respond with greater speed and intensity to P. caspici?; 2) Are there R proteins or other receptors, not described here, involved in the process?; 3) Is this rapid response linked to the differential expression of the genes of the mevalonic path HMG1, HMG2, HMG3, EAS and to the increased activity of the octodecaoinc path in resistant cultivars?; 4) Could the application of ACBs be a strategy to increase tolerance levels in susceptible cultivars? And if so, how long will the protection last?
Final considerations. The critical analysis of the state of the art of this pathosystem helped discern that the specific resistance of Capsicum to P. capsici is the result of a process coordinated by growth regulators JA, ET and SA. This entails an exception to the antagonism commonly mentioned between these signaling paths. However, further studies are required that take over the role of each growth regulator in the process of signaling in the defense against this oomycete. Although it is clear that genes such as PAL, HMG, EAS, GLU, CHI and POX are broadly related with the process of defense and resistance of Capsicum to P. capsici, underneath there is a large number of genes that are positively and negatively affected by the paths of signaling of these growth regulators. Because this is a phenomenon with multifactorial causes, the new massive data generation technologies may offer a wider outlook regarding activated and suppressed genes during interaction. It is also important to highlight the implication of these growth regulators in the process of resistance induction by beneficial microorganisms, which are an important component of the soil native microbiota that establish associations with the pathosystem. The microbial diversity plays an important role, since it directly affects the result of the molecular dialogue; first, since it acts as a barrier to complicate or prevent the phytopathogen from establishing on the plant, and secondly, by increasing the defensive ability by activating the basal defense and inducing resistance. Despite the progress made, there are still many questions to be answered. As the comprehension of pathosystems advances, and as investigation covers the complexity of agricultural ecosystems, the impact of phytopathogens can gradually be reduced. The control of plant pathogens should not be directed to their exclusion from the agroecosystems, but rather, to employ ecological technologies that promote the balance of the populations of soil microorganisms, which eventually will allow to achieve a sustainable management of plant pathogens and pests.
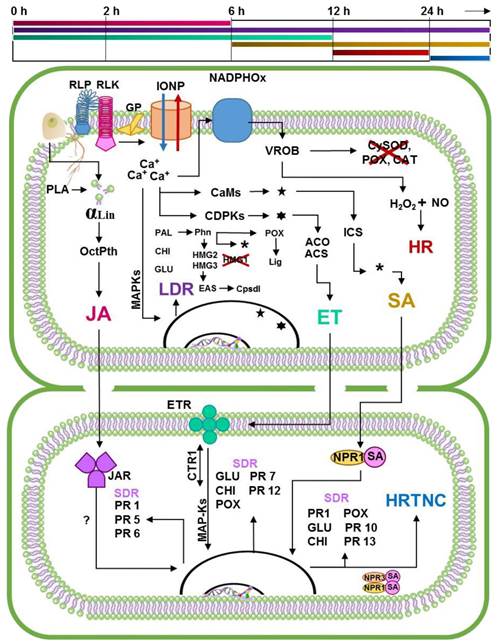
Figure 3 Proposal of the molecular dialogue that takes place in the
specific resistance of Capsicum to
Phytophthora capsici. In the upper part of the
figure appears a line that marks the times in which the events
related to specific resistance from Capsicum to
P. capsici are estimated to occur. The first
line corresponds to the JA signaling pathway. The second correspond
to local defense responses activated in each infection point, which
occurs in similarly to the MTI/PTI process. Third line correspond to
the ET signaling pathway. Fourth line indicates the SA signaling
pathway. The fifth line corresponds to the activation of local HR
induced by ROs in resistant genotypes, while the sixth indicates the
activation of HR in neighbor cells induced by the constant increase
of SA concentration. Based on the critical analysis of the state of
the art of the pathosystem in question, this proposal suggests that
the specific resistance of Capsicum to P.
capsici is the result of the summation of a series of
coordinated events, in which the main actors are the plant growth
regulators involved in the signaling pathways (Egea et
al., 1996; Kim y Hwang, 2000; Jung et
al., 2005; Lee y Hawng, 2005; Bent y Makey, 2007;
Conrath, 2009; Ozgonen et al., 2009; Ueeda
et al., 2006; Yi et al., 2010;
Glowacki et al., 2011; Jingyuan et
al., 2011; López-Martínez et al.,
2011; Sanzón-Gómez y Zavaleta-Mejía, 2011; Bertoni, 2012; Gururani
et al., 2012; Sudisha et al.,
2012; Castro-Rocha et al., 2012; Muthamilarasan y
Pasard, 2013; Veloso et al., 2014; Villar-Luna
et al., 2015; Vidhyasekaran, 2014; Villar-Luna
et al., 2009, 2017). αLin Linoleic acid; ACS
ACC Syntase; ACO ACC Oxidase; CaMs Calmodulines; CDPKs Calcium
dependent kinases; CHI PR3-Chitinases; CTR1 Raf kinase considered as
a MAP-K; Cpsdl Capsidiol synthesis; CySOD Superoxide dismustase in
the capsidiol; EAS 5-epi-aristolochene synthase; ET Ethylene; ETR
Ethylene receptors; GLU PR2-Glucanases; GP G proteins ; HMG 1, 2, 3
family of genes that codifies hidoxymethylglutaril-coenzyme A
reductase; HR Hypersensitive Response; HRTNC HR Triggered in
Neighbor Cells; ICS Isochorismate Synthase; IONP Ion Pumps; JAR JA
receptors; JA Jasmonic acid; LDR Local Defense Responses; Lig Cell
wall lignification process; RLK Receptor-Like Kinases; RLP
Receptor-like Protein; MAPKs Mitogen Activated Protein Kinases;
NADPHOx NADPH oxidase; NO Nitric oxide; NPR 1, 3 Receptors with high
affinity to SA; OctPth Octadecanoid pathway; PAL Phenilalanine
Ammonia Lyase; PLA Phospholipase A; Phn Phenols synthesis; PR1
Pathogenesis related protein from Group 1; PR5 Thaumatin; PR6
Protease inhibitor; PR7 Endoproteinases; PR12 Defensins; POX
PR9-Peroxidases; SA Salicylic acid; VROB Violent ROs Burst;
*, ,
, , Goes
to.
, Goes
to.
Acknowledgements
To the National Science and Technology Council - CONACyT for the scholarship granted to the main author.
REFERENCES
Bae H, Roberts DP, Lim HS, Strem MD, Park SC, Ryu CM, Meinick RL, Bailey BA. (2011). Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against P. capsici in hot pepper using multiple mechanisms. Mol. Plant-Microbe Interac. 24:336-351. Doi: https://doi.org/10.1094/MPMI-09-10-0221 [ Links ]
Barchenger DW, Lamour KH, Bosland PW. (2018). Challenges and Strategies for Breeding Resistance in Capsicum annuum to the Multifarious Pathogen, Phytophthora capsici. Front. Plant Sci. 9:628. Doi: http://journal.frontiersin.org/article/10.3389/fpls.2018.00628/full [ Links ]
Bent AF, Makey D. (2007). Elicitors, Effectors and R genes: The new paradigm and lifetime supply of questions. Annu. Rev. Phytopathol. 45:399-436. Doi: https://doi.org/10.1146/annurev.phyto.45.062806.094427 [ Links ]
Bertoni G. (2012). Oxylipins and plant palatability. Plant Cell. 24: 1305. Doi: https://doi.org/10.1105/tpc.112.240412 [ Links ]
Bishop HSL, Mounter SA, Laskey J, Morris RO, Elder J, Roop P, Rouse C, Schmidt FJ, English JT. (2002). Phage-Displayed peptides as developmental agonists for Phytophthora capsici zoospores. App.Envir. Microbiol. 68:3315-3320. Doi: https://doi.org/10.1128/AEM.68.7.3315-3320.2002 [ Links ]
Bostock RM, Saychenko C, Lazarus C, Dehesh K. (2011). Eicosapolyenoic acids. Novel MAMPs with reciprocal effect on oomycete-plant defense signaling networks. Plant Signal. Behav. 6:531-533. Doi: https://doi.org/10.4161/psb.6.4.14782 [ Links ]
Brich PRJ, Boevink PC, Gilroy EM, Hein I, Pritchard L, Whisson SC. (2008). Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11:337-379. Doi: https://doi.org/10.1016/j.pbi.2008.04.005 [ Links ]
Cannon SB, Zhu H, Baumgarte AM, Spangler R, May G, Cook DR, Young ND. (2002). Diversity, distribution and ancient taxonomic relationship within the TIR and Non-TIR NBS-LRR resistance gene subfamilies. J. Mol. Evol. 54:548-562. Doi: https://doi.org/10.1007/s0023901-0057-2 [ Links ]
Castro-Rocha A, Fernández-Pavia SP, Osuna-Avila P. (2012) Chili defense mechanisms in the Capsicum annuum-Phytophthora capsici pathosystem. R.M.F. 30:49-65. En línea: http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S0185-33092012000100005 [ Links ]
Chemeltorit PP, Mutaqin KH, Widodo W. (2017). Combining Trichoderma hamatum THSW13 and Pseudomonas aeruginosa BJ10-86:a synergistic chili pepper seed treatment for Phytophthora capsici infested soil. Eur. J. Plant. Pathol. 147:157-166. Doi: https://doi.org/10.1007/s10658-016-0988-5 [ Links ]
Chen YY, Chen PC, Tsay TT. (2016). The biocontrol efficacy and antibiotic activity of Streptomyces plicatus on the oomycete Phytophthora capsici. Bio. Con. 98:34-42. Doi: https://doi.org/10.1016/j.biocontrol.2016.02.011 [ Links ]
Conrath U. (2009). Priming of induced plant defense responses. Adv. Bot. Res. 51:361-395. Doi: https://doi.org/10.1016/S0065-2296(09)51009-9 [ Links ]
Dahiya N, Tewari R, Hoondal GS. (2006). Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71:773-782. Doi: https://doi.org/10.1007/s00253-005-0183-7 [ Links ]
Egea C, Alcázar MD, Candela E. (1996). Capsidiol: Its role in the resistance of Capsicum annuum to P. capsici. Physiol. Plantarum. 98:737:742. Doi: https://doi.org/10.1111/j.1399-3054.1996.tb06679.x [ Links ]
Feng B, Li P, Wang H, Zhang X. (2010). Functional analysis of Pcpme6 fom oomycete plant pathogen Phytophthora capsici. Microb. Pathog. 49:23-31. Doi: https://doi.org/10.1016/j.micpath.2010.03.004 [ Links ]
Fernández-Herrera E, Rojas-Martínez RI, Gómez-Rodríguez O, Guevara-Olvera L, Rivas-Dávila ME, Valdez-Moctesuma E, Zavaleta-Mejía E. (2012). Genes de defensa, actividad enzimática y contenido de capsidiol en chile CM-334 inoculado con Phytophthora capsici. Interciencia. 37:370-377. En línea: http://www.redalyc.org/articulo.oa?id=33922756007 [ Links ]
French MRD, Jones JB, Ozores HM, Roberts PD. (2007). Survival of inoculum of Phytophthora capsici in soil trough time under different soil treatment. Plant Dis. 91:593-598. Doi: https://doi.org/10.1094/PDIS-91-5-0593 [ Links ]
Glowacki S, Macoiszek VK, Kononowicz AK. (2011). R proteins as fundamentals of plant innate immunity. Cell. Mol. Biol. Letters. 16:1-24. Doi: https://doi.org/10.2478/s11658-010-0024-2 [ Links ]
Goldberg NP. (2001) Chile Pepper Diseases Circular 549. College of Agriculture, Consumer and Environmental Science. New Mexico State Universiy. En línea: https://aces.nmsu.edu/pubs/_circulars/CR549/welcome.html [ Links ]
Gururani MA, Vankatesh J, Upadhyaya CP, Nookaraju A, Padney SK, Park SW. (2012). Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 78:51-65. Doi: https://doi.org/10.1016/j.pmpp.2012.01.002 [ Links ]
Hardham AR, Shan W. (2009). Cellular and molecular biology of Phytophthora-Plant Interactions. In: Deising H (Ed.) Plant Relationships, 2nd Edition The Mycota V. Edition. Springer-Verlag Berlin. pp. 3-27. Doi: https://doi.org/10.1007/978-3-540-87407-2_1 [ Links ]
Jiménez-Camargo A, Valadez-Moctezuma E, Lozoya-Saldaña H. (2018). Antagonism by Penicillum sp. Against Phytophthora capsici (Leonian). Rev. Fitotec. Mex. 41:137-148. En línea: https://www.revistafitotecniamexicana.org/documentos/41-2/5a.pdf [ Links ]
Jingyuan Z, Xuexiao Z, Zhenchuan M, Bingyan X. (2011). A novel pepper (Capsicum annuum L.) WRKY Gene CaWRKY30, is involved in pathogen stress responses. J. Plant Biol. 54:329-337. Doi: https://doi.org/10.1007/s12374-011-9171-x [ Links ]
Jung WJ, Jin YL, Kim KY, Park RD, Kim TH. (2005). Changes in pathogenesis-related proteins in pepper plants with regard to biological control of phytopthora blight with Paenibacillus illinoisensis. BioControl. 50:165-178. Doi: https://doi.org/10.1007/s10526-004-0451-y [ Links ]
Jupe J, Stam R, Howden AJM, Morrin JA, Zhang R, Hedley PE, Huitema E. (2013). Phytophthora capsici-tomato interaction features dramatic shifts gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 14:R63. Doi: https://doi.org/10.1186/gb-2013-14-6-r63 [ Links ]
Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Physiol. 44:41-60. Doi: https://doi.org/10.1146/annurev.phyto.44.070505.143436 [ Links ]
Kim BS, Lee JY, Hwang BK. (2000). In vivo control and in vitro antifungal activiry of rhamnolipid B, a glycolipid antibiotic, against Phytophthora capsici and Colletotrichum orbiculare. 56:1029-1035. Doi: https://doi.org/10.1002/1526-4998(200012)56:12<1029::AID-PS238>3.0.CO;2-Q [ Links ]
Lamour KH, Stam R, Jupe J, Huitema E. (2012). The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 13:329-337. Doi: https://doi.org/10.1111/j.1364-3703.2011.00754.x [ Links ]
Lee JH, Hong JP, Oh SK, Lee S, Choi D, Kim WT. (2004). The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequence with different binding affinities possible biological roles of CaERFPL1 in response to pathogen interactions and high salinity conditions in transgenic tobacco plants. Plant. Mol. Biol. 42:335-344. Doi: https://doi.org/10.1007/s11103-004-0417-6 [ Links ]
Lee SC, Hwang BK. (2005). Induction of some defense-related genes and oxidative burst is required for the establishment of systemic acquired resistance in Capsicum annuum. Planta. 221: 790-800. Doi: https://doi.org/10.1007/s00425-005-1488-6 [ Links ]
Ley-López N, Márquez-Zequera I, Carrillo-Fasio JA, León-Félix J, Cruz-Lachica I, García-Estrada RS. (2018). Effect of biocontrol and germinative inhibition of Bacillus spp. On zoospores of Phytophthora capsici. RMF. 36:215-232. Doi: http://dx.doi.org/10.18781/R.MEX.FIT.1711-2 [ Links ]
López-Martínez NM, Colinas-León T, Peña-Valdivia CB, Salinas-Moreno Y, Fuentes-Montiel P, Biesaga M, Zavaleta-Mejía E. (2011). Alterations in peroxidase activity and phenylpropanoid metabolism induced by Nacobbus aberrans Thorne and Allen in chilli (Capsicum annuum L.) CM334 resistant to Phytophthora capsici Leo. Plant and soil. 338:399-409. Doi: https://doi.org/10.1007/s11104-010-0553-5 [ Links ]
Minamiyama Y, Tsuro M, Kubo T, Hirai M. (2005). QTL analysis for recistance to Phytophtthora capsici in pepper using a high density SSR-based map. Breed. Sci. 57:129-134. Doi: https://doi.org/10.1270/jsbbs.57.129 [ Links ]
Mongkolporn O, Taylor PWJ. (2011). Capsicum. In: Wild Crop Relatives: Genomics and Breeding Resourses, Vegetables. (Ed) Kole, C. Springer-Verlag Berlin Heidelberg. Pp. 43-57. Doi: https://doi.org/10.1007/978-3-642-20450-0 [ Links ]
Muthamilarasan M, Prasad M. (2013). Plant innate immunity: An updated insight into defense mechanism. J. Biosci. 38:1-17. Doi: https://doi.org/10.1007/s12038-013-9302-2 [ Links ]
Nespoulous C, Gaudemer O, Huet JC, Pernollet JC. (1999). Characterization of elicitin-like phospholipases isolated from Phytophthora caspici culture filtrate. FEBS letters. 452:400-406. Doi: https://doi.org/10.1016/S0014-5793(99)00654-7 [ Links ]
Ozgonen H, Yardimci N, Kilic HC. (2009). Induction of phenolic compounds and pathogenesis-related proteins by mycorrhizal fungal inoculations against Phytophthora capsici Leonian in pepper. Pak. J. Biol. Sci. 12:1181-1187. Doi: 10.3923/pjbs.2009.1181.1187. En línea: https://scialert.net/abstract/?doi=pjbs.2009.1181.1187 [ Links ]
Ramos SRU, Gutiérrez SJG, Rodrígez GR, Salcedo MSM, Hernández LCE, Luna OHA, Jiménez BJF, Fraire VS, Almeyda LIH. (2010) Antagonismo de dos ascomicetos contra Phytophthora capsici Leonian, causante de la marchitez del chile (Capsicum annuum L.) RMF. 28:75-86. En línea: http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S0185-33092010000200001 [ Links ]
Reifschneider FJB, Boiteux LS, Della Vecchia PT. (1992). Inheritance of resisance in peppers to Phytophthora capsici in pepper. Euphytica 62:45-49. https://doi.org/10.1007/BF00036086 [ Links ]
Sanzón-Gómez D, Zavaleta-Mejía E. (2011). Respuesta de hipersensibilidad, una muerte programada para defenderse del ataque por fitopatógeno. RMF. 29:154-164. En línea: http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S018533092011000200007&lng=es&nrm=iso [ Links ]
Stam R, Jupe J, Howden AJM, Morris JA, Bovenik PC, Heldey PE, Huitema E. (2013). Identification and characterization CRN effectors in Phytophthora capsici show modularity and functional diversity. PLoS ONE 8(3):e59517. En línea: http://www.plosone.org/article/citationList.action?articleURI=info%3Adoi%2F10.1371%2Fjournal.pone.0059517 [ Links ]
Sudisha J, Sarathchandra RG, Amrutesh KN, Kumar A, Shetty HS (2012). Pathogenesis Related Proteins in Plant Defense Response. In: Plant Defence: Biological Control. Progress in Biological Control Vol 12. (Eds) Mérillon J, Ramawat K. ED Springer Dordrecht. Pp. 379-403. Doi: https://doi.org/10.1007/978-94-007-1933-0_17 [ Links ]
Thomma BPHJ, Nürnebereger T, Joosten MHAJ. (2011). Of PAMPs and effectors: The blurred PTI-ETI Dichotomy. The Plant Cell. 23:4-15. Doi: https://doi.org/10.1105/tpc.110.082602 [ Links ]
Tian MY, Win .J, Song J, van Der HR, van Der KE, Kamoun S. (2016). A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 143:364-377. Doi: https://doi.org/10.1104/pp.106.090050 [ Links ]
Tör M. (2008). Tapping into molecular conversation between oomycete plant pathogens and their hosts. Eur. J. Plant Pathol. 122:57-69. Doi: https://doi.org/10.1007/s10658-008-9288-z [ Links ]
Torto AT, Collmer CW, Lindeber M, Bird D, Collmer A, Tyler BM. (2009). Common and contrasting themes in host cell-targeted effectors from bacterial, fungal, oomycete and nematode plant symbionts described using the Gene Ontology. BMC Microbiol. 9:1-8. Doi: https://doi.org/10.1186/1471-2180-9-S1-S3 [ Links ]
Ueeda M, Kubota M, Nishi K. (2006). Contribution of jasmonic acid to resistance against Phytophthora blight in Capsicum annuum cv. SCM334. Physiol. Mol. Plant. Pathol. 67:149-154. Doi: https://doi.org/10.1016/j.pmpp.2005.12.002 [ Links ]
Veloso J, Díaz J. (2012). Fusarium oxysporum Fo47 confers protection Pepper plants against Verticillium dahlia and Phytophthora capsici, and induces the expression of defence genes. Plant Pathol. 61:281-188. Doi: https://doi.org/10.1111/j.1365-3059.2011.02516.x [ Links ]
Veloso J, García T, Bernal A, Díaz J. (2014). New bricks on the wall of induced resistance: salicylic acid receptors and transgenerational priming. Eur. J. Plant Pathol. 38:685-693. Doi: https://doi.org/10.1007/s10658-013-0350-0 [ Links ]
Vidhyasekaran P. (2014). PAMP signaling in plant innate immunity: Signal perception and transduction. In: Signaling and communication in plants series number 21. Springer Science+Busines Media Dordrecht. Pp. 17-161. Doi: https://doi.org/10.1007/978-94-007-7426-1 [ Links ]
Villar-Luna E, Reyes-Trejo B, Rojas-Martínez RI, Gómez-Rodríguez O, Hernández-Anguiano AM, Zavaleta-Mejía E. (2009). Respuesta hipersensitiva en follaje de chile CM-334 resistente a Phytophthora capsici infectado con Nacobus aberrans. Nematropica. 39:143-155. En línea: http://journals.fcla.edu/nematropica/article/viewFile/64475/62143 [ Links ]
Villar-Luna E, Rojas-Martinez R, Reyes-Trejo B, Gómez-Rodríguez O, Zavaleta-Mejía E. (2017). Mevalonate pathway genes expressed in chilli CM334 inoculated with Phytophthora capsici and infected by Nacobbus aberrans and Meloidogyne enterolobii. Eur. J. Plant Pathol. 148:867-881. Doi: https://doi.org/10.1007/s10658-016-1142-0 [ Links ]
Villar-Luna H, Reyes-Trejo B, Gómez-Rodríguez O, Villar-Luna E, Zavaleta-Mejía E. (2015) Expresión de genes de defensa y acumulación de capsidiol en la interacción compatible CM334/ Nacobus aberrans e incompatible CM334/Meloidogyne incognita. Nematropica 45:9-19. En línea: http://journals.fcla.edu/nematropica/article/view/85048/81977 [ Links ]
Win J, Chaparro GA, Belhaj K, Saunders DG, Yoshida K, Dong S, Shornack S, Zipfel C, Robatzek S, Hogenhout SA, Kamoun S. (2012). Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb Symp Quant. Biol. 77:235-47. Doi: http://dx.doi.org/10.1101/sqb.2012.77.015933 [ Links ]
Xu S, Kim BS. (2016). Evaluation of Paenibacillus polymyxa strain SC09-21 for biocontrol of Phytophthora blight and growth stimulation in pepper plants. Trop. Plant Pathol. 41:62. Doi: https://doi.org/10.1007/s40858-016-0077-5 [ Links ]
Yi SY, Lee DJ, Yeom SI, Yoon J, Kim YH, Kwon SY, Choi D. (2010). A novel pepper (Capsicum annuum) receptor-like kinase functions as a negative regulator of plant cell death via accumulation of superoxide anions. New Phytol. 185:701-705. Doi: https://doi.org/10.1111/j.1469-8137.2009.03095.x [ Links ]
Received: January 17, 2019; Accepted: March 05, 2019











 texto em
texto em 


