Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.37 n.1 Texcoco Jan. 2019 Epub Aug 21, 2020
https://doi.org/10.18781/r.mex.fit.1809-3
Scientific articles
Response of Capsicum pubescens genotypes to damage caused by the fungal wilt complex
1 Centro Universitario Tenancingo, Universidad Autónoma del Estado de México, Km 1.5. Carretera Tenancingo-Villa Guerrero, Tenancingo Estado de México, C.P. 52400, México;
2 División de Ingeniería en Innovación Agrícola Sustentable, Tecnológico de Estudios Superiores de Villa Guerrero, Carretera Federal México-Ixtapan de la Sal km 64.5. La Finca, Villa Guerrero, Estado de México. C.P. 51760, México.
In the south of the State of Mexico the manzano pepper (Capsicum pubescens R. and P.) is an economically important crop, but is affected by the “wilt disease”, a root disease that causes the death of the plant. The objective was to identify the associated organisms and evaluate the variation in response to the damage of wilt using 16 genotypes (M1-M16) of manzano pepper. Segments of infected plants were sown in PDA and 3P culture medium. The associated organisms were morphologically and molecularly identified. To evaluate the pathogen-genotype interaction, associated organisms were inoculated alone and their combinations in C. pubescens. Fusarium oxysporum, Phytophthora capsici and Rhizoctonia solani, were identified as the main cause of wilt, in which there were differences in severity and incidence between colonies of F. oxysporum (P≤0.01), and P. capsici (P≤0.05). There were significant differences (P≤0.01) in pathogenicity between P. capsici, F. oxysporum and R. solani, and combinations among them. Variation in resistance to wilt was observed, where M8 was the genotype that showed resistance to F. oxysporum and R. solani, and M9 tolerance to F. oxysporum. These can be used in breeding for the development of resistant genotypes.
Key words: Manzano pepper; Phytophthora capsici; Rhizoctonia solani; Fusarium oxysporum
En el sur del Estado de México el chile manzano (Capsicum pubescens R. y P.) es un cultivo económicamente importante, sin embargo, es afectado por la “marchitez”, enfermedad de raíz que provoca la muerte de la planta. El objetivo fue identificar los organismos asociados y evaluar la variación en respuesta al daño de la marchitez en 16 genotipos (M1-M16) de chile manzano. Se sembraron segmentos de plantas infectadas en medio de cultivo PDA y 3P y se identificaron morfológica y molecularmente a los organismos asociados. Para la interacción patógeno-genotipo, se inocularon los organismos asociados solos y sus combinaciones en C. pubescens. Se identificaron a Fusarium oxysporum, Phytophthora capsici y Rhizoctonia solani como responsables de la marchitez, en los que hubo diferencias en severidad e incidencia entre colonias de F. oxysporum (P≤0.01), y de P. capsici (P≤0.05). Hubo diferencias significativas (P≤0.01) en patogenicidad entre P. capsici, F. oxysporum y R. solani, y combinaciones entre estos. Se observó variación en la resistencia a la marchitez, donde M8 fue el genotipo que presentó resistencia a F. oxysporum y R. solani, y M9 tolerancia a F. oxysporum. Éstos pueden ser aprovechados en el mejoramiento genético para desarrollo de genotipos resistentes.
Palabras clave: chile manzano; Phytophthora capsici; Rhizoctonia solani; Fusarium oxysporum
The Capsicum genus, which includes sweet and hot chilies, are vegetables and spices cultivated and consumed worldwide (Carrizo et al., 2016). According to USDA-ARS (2011), the genus Capsicum has 38 species, from which the most grown are Capsicum annuum, Capsicum frutescens, Capsicum chinense, Capsicum pubescens and Capsicum baccatum. From the five species, C. annuum and C. frutescens were domesticated in Mesoamerica, and C. chinense, C. baccatum and C. pubescens in South America (Pickersgill, 2007). Traditionally and historically, chili is an important crop in Mexico, whose production in 2016 was of 2.3 million tons valued at more than 22,500 million Mexican pesos (SAGARPA, 2017). Among the diversity of chili species cultivated in Mexico, the importance of manzano pepper (C. pubescens) increased in the last decade, and according to SAGARPA (2018), 4,221.83 tons are intended only for export to the United States, 86.3% of which are produced in the southern zone of the State of Mexico. Manzano pepper cultivation is intensive in that region. However, manzano pepper has agronomic production limitations, such as wilt susceptibility, a disease that causes root damage and death in manzano pepper plants. This disease, which is considered the most destructive worldwide (Zhang et al., 2013), was first detected in C. annuum by Leonian (1922), who identified Phytophthora capsici as the causal agent. The associated organisms affect the crop at any development stage, cause loss of turgidity and the subsequent death of plants (Kousik et al., 2012). Besides P. capsici, in Mexico, Rhizoctonia solani and Fusarium oxysporum have been reported as causal agents of wilt in C. annuum (Uc-Arguelles et al., 2017).
According to González-Pérez et al. (2014), C. pubescens has low genetic diversity as a result of a founder effect during its domestication that makes it a less polymorphic species. However, in the southern zone of the State of Mexico, with a varied number of ecologic niches, different genotypes of C. pubecens are cultivated, which represents a considerable variation within the species and, therefore, a valuable germplasm reservoir that could be used to improve not only the morphological traits but also the disease resistance of this species (Arias et al., 2017). With the existing natural variation, some individuals from a population can express certain characteristics with a higher or lesser degree than others, and this can give the individual certain advantages linked to their life cycle. Fruits produced by those individuals may vary both in number and size, color, texture, taste, maturity, appearance and quality, as well as plant’s architecture and ability to resist abiotic or biotic stresses (Schubert et al., 2009). Due to these factors, the variation in the reproduction of C. pubescens genotypes, during which some individuals may have a higher number of offspring than those of others, would mean that there is an increase in the frequency of their genetic material compared to others (Nora et al., 2011).
The genetic diversity within species is the main reason why a determined species evolves under changing environmental conditions and selection pressures. Knowing the genetic diversity is key in order to diversify germplasm sources, try to minimize genetic vulnerability risks and increase the probabilities of detecting favorable alleles (Ruíz et al., 2016). The University Center of the Autonomous University of Mexico State (UAEM for its acronym in Spanish) in Tenancingo has 16 genotypes that were collected in the southern zone of the State of Mexico, 15 of which represent morphological variability and have unique traits that are genetically and commercially important (Martínez, 2016). To make the most of the C. pubescens genotypes that are part of the collection of the UC-Tenancingo, the objective of the present study was to evaluate the response of 16 manzano pepper genotypes to the fungal complex of microorganisms associated with chili wilt in the State of Mexico.
Materials and methods
The research was conducted at UAEM’s University Center in Tenancingo, located at 18° 58’ 05.53’’ N and 99° 36’ 50.51’’ W, 2068 masl. In 2016, manzano pepper plants showing wilt symptoms were collected at the following locations of the southern zone of the State of Mexico: 5 in Ahuacatitlán, Ixtapan de la Sal; 5 in El Zarco, Tenancingo; 3 in Santa Ana, Tenancingo; 4 in Tepoxtepec, Tenancingo; 3 in Matlazinca, Villa Guerrero; 4 in San Miguel, Ixtapan de la Sal; 5 in El Potrero, Coatepec Harinas; 5 in Ixtlahuaca, Coatepec Harinas; 3 in San Nicolás, Tenancingo; and 4 in Las Cabañas, Tenancingo. The collection sites were classified according to their cropping intensity (IC) that was defined based on the cultivated area: low: up to 3 ha; intermediate: 3.1-8.0 ha; high: more than 8.1 ha. Samples wrapped with wet brown paper were placed in transparent plastic bags and transported in a cooler to the laboratory, where were kept at 4 ºC until the next day, when they were used.
Isolation of the associated organisms. Eight segments of root and eight segments of stem were taken from each of the 41 plants collected in the 10 sampled sites, with a total of 656 samples. The samples were disinfected; for fungi, the samples were placed in Petri dishes containing a potato-dextrose-agar (PDA) culture medium, according to the method of López (1984); oomycetes were cultivated in 20 g of maize flour, 18 g of agar-agar, 0.8 mL of pimaricin, 0.02 g of rifamycin and 0.25 g L-1 of ampicillin diluted in distilled water, according to the method of López et al., (2009), modified from Kannwischer and Mitchell (1978). The cultures were incubated at 24 oC in darkness and monitored every 24 h to observe fungus growth and development. When fungi and oomycetes had grown, successive transfers were made to obtain pure cultures and then prepare monosporic and hyphal tip cultures. The colonies were kept in tubes containing a PDA culture medium covered with sterile mineral oil.
Morphological identification of the associated organisms. The associated organisms were identified using taxonomic keys for fungi and oomycetes. The keys of Booth (1971) and Leslie and Summerell (2006) were used to characterize fungi micronidia and macronidia size and shape, and to describe chlamydospores, which are an important structure for identification; and the keys of Singlenton et al. (1992) and Watanabe (2002) to characterize sclerotia, ramification angles and mycelium. The keys of Erwin and Ribeiro (1996) and Gallegly and Hong (2008) were used to identify oomycetes colony growth, type of mycelium, and sporangia and chlamydospores shape.
Molecular identification of the associated organisms. DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method and sodium acetate. Universal PCR reactions were performed for fungi and oomycetes using the ITS-1 5’-tccgtaggtgaacctgcgg-3’ and ITS-4 5’-tcctccgcttattgatatgc-3’ primers (White et al., 1990) that amplify fragments of 500-900 base pairs (bp). The reaction mixture in a final volume of 25 μL was made from a mixture of 10 µL of ultrapure water, 12 µL of MyTaq Mix® (Bioline), 1 µL of F primer, 1 µL of R primer and 1 µL of DNA. For oomycetes, the thermal program was set at 94 °C for 2 min, followed by 35 cycles at 94-55-72 °C for 30-30-60 s, respectively, and one final extension at 72 °C for 5 min. For fungi, the thermal program was set at 94 °C for 2 min, followed by 35 cycles at 94-60-72 °C for 30-30-60 s, respectively, and one final extension at 72 °C for 5 min. The amplified products from the PCR reactions were separated by electrophoresis using tris-borate-EDTA (TBE) and 1.2% agarose gels.
Sequencing. The PCR amplified fragments were purified using the EZ-10 Spin Column Handbook kit (Bio Basic Canada, Inc.) and then sequenced in the Molecular Biology Laboratory of FES-Iztacala, Universidad Nacional Autónoma de México (UNAM). The sequences in FASTA format (Chromas 2.6.5 version) were aligned with the National Center for Biotechnology Information (NCBI) gene bank database for consensus and identity -BLAST (Basic Local Alignment Search Tool) (available at https://www.yeastgenome.org/blast-fungal).
Pathogenicity in vitro. Pathogenicity was evaluated on C. pubescens M3 genotype, the most cultivated material in the study region. Seeds were disinfested and inoculated following the method of Apodaca-Sánchez et al. (2001), and then placed in Petri dishes containing an agar-water culture medium. Each strain obtained from the monosporic and hyphal tip cultures was considered as a treatment of 5 repetitions with 10 seeds each. Disinfested seed immersed in sterile distilled water was used as control. The pathogenicity was evaluated using the incidence and severity levels measured at day 10, according to Herrera and Laurentin (2012), and radicle and hypocotyl symptoms were recorded. The incidence was determined using the percentage of seedlings showing disease symptoms. The severity was measured using the lesion length along the seedling and expressed as percentage. The obtained values were averaged within each experiment unit. The ratio between incidence and severity was calculated proportionally using the severity/incidence quotient (Segura et al., 2009; Fernández et al., 2010).
Interaction in multiple infections. Disinfested seeds of the M3 genotype were placed in a test tube containing an agar-water medium, kept in darkness at 25 °C, and, when germinated, at intervals of 12 h light and 12 h darkness at 25 °C. At day 7, the seeds were inoculated following the method of Herrera and Laurentin (2012). A 100 µL of 1 x 106 spore suspension and 100 µL of monosporic and hyphal tip culture of the associated organisms, alone and combined, were applied to seven treatments with 10 replications each: P. capsici (P); P. capsici plus R. solani (P+R); F. oxysporum plus P. capsici (F+P); F. oxysporum plus P. capsici plus R. solani (F+P+R); F. oxysporum (F); and R. solani (R). The treatments were incubated at intervals of 12 h light and 12 h darkness at 25 °C and monitored every 24 h to evaluate the disease severity, which was determined by lesion length/seedling length x 100.
Pathogens-genotype interaction. C. pubescens M1-M16 genotypes collected at different sites of the southern zone of the State of Mexico were used because they are morphologically contrasting with different traits such as flower and fruit (Martínez, 2016). Seedlings with four true leaves were inoculated with the identified pathogens, alone and combined, according to the following treatments (T): F. oxysporum (T1); P. capsici (T2); R. solani (T3); F. oxysporum + P. capsici (T4) and F. oxysporum + P. capsici + R. solani (T5). The inoculation method used was that described by Martínez et al. (1996). The plants inoculated were transplanted to 1-liter polystyrene cups containing peat and expanded mineral perlite (agrolita) at a 2:1 ratio, respectively, with five replications per treatment each. The disease severity was measured by the percentage of the seedling’s hypocotyl and root using a 1-9 scale developed at CIAT (Abawi and Pastor-Corrales, 1990). Data were logarithmically converted to obtain a general linear model using the InfoStat statistical software (Di Rienzo et al., 2016).
Statistical analysis. The data obtained were subjected to an analysis of variance and the treatments were compared using Duncan’s test (p=0.05) and the InfoStat program.
Results and discussion
Isolation of the associated organisms. Fusarium oxysporum, Phytophthora capsici and Rhizoctonia solani (Table 1) were isolated from samples collected at the study sites. F. oxysporum was constantly found in all the sites (9/10), while P. capsici was found only in sites with intermediate and high cropping intensity (Table 1). The presence of R. solani in one sampled site suggests a limited participation in wilt damage. The three associated organisms were isolated at one site, and this indicates that the pathogen may individually or collectively affect the host. These results coincide with those reported by Anaya-López et al. (2011), who mentioned F. oxysporum as the most common pathogen, followed by R. solani in C. annuum crops. According to Nelson et al. (1983), the frequency variation of the associated organisms that cause wilt is affected by diverse factors, among which climate conditions are crucial. Compared to the other communities, the climate at El Potrero, the site where the three associated organisms were found, is warmer and rainy, and favors the development of the associated organisms. On the other hand, the crop systems, the use of fungicides and the genotype of the same crop itself, are decisive factors that favor the presence and survival of the organisms associated with the fungal complex (Guigón-López et al., 2001; Lozano et al., 2015). The crop’s phenological stage at which samples are collected could influence the presence of the fungal complex, as has been observed in the case of C. annuum seedling stage (Vásquez et al., 2009).
Morphological description of the associated microorganisms identified. Fusarium oxysporum developed abundant aerial, hyaline and septate mycelium, with radial growth habit and orange pigmentation, oval microconidia of 8.6 x 4.3 µm average size with 0 or 1 septum, abundant in fake heads and in monophialides, a lower number of macroconidia with attenuated apical cell and foot-like basal cell with 3-4 septa of 23.8 x 4.2 µm average size, as well as abundant intercalary and terminal chlamydospores on hyphae (Figure 1).
Table 1. Isolated organisms from C. pubescens crops at three municipalities in the southern zone of the State of Mexico.
| Localidad | Microorganismo fitopatógeno | Muestras procesadasy | |||
|---|---|---|---|---|---|
| Zona | ICz | F.oxysporum | P. capsici | R. solani | |
| Ahuacatitlan | Baja | X | 75 | ||
| El Zarco | Media | X | X | 75 | |
| Santa Ana | Baja | X | 45 | ||
| Tepoxtepec | Baja | X | 60 | ||
| Matlazinca | Media | X | X | 45 | |
| San Miguel | Media | X | X | 60 | |
| El Potrero | Alta | X | X | X | 75 |
| Ixtlahuaca | Alta | X | X | 75 | |
| San Nicolás | Baja | X | 45 | ||
| Las Cabañas | Media | X | 60 | ||
| Total | 656 | ||||
z Intensidad de cultivo; ysegmentos de tallo y raíz / z Cropping intensity; ystem and root segments.
The first four days, Rhizoctonia solani developed mycelium white in color, which turned brown at day five, and sclerotia. Microscopical observations showed young vegetative cells with a ramification next to the distal septum, a 90° angle, narrow hyphae and formation of septa at a short distance from the point of origin to the hyphal ramification, 5-8 μm wide hyphae and sclerotia 1-3 mm in diameter (Figure 1).
Phytophthora capsici developed mycelium white in color, rosette-like growth in PDA culture medium, and coenocytic, hyaline and torulose mycelium; ovoid, lemon-like and not fully papillated sporangia of 11 a 54 µm long and 9 a 36 µm wide, and intercalary chlamydospores of 9.5 µm in diameter (Figure 1).
Different authors (Lozano et al., 2015) have reported the associated organisms as the causal agents of wilt in C. annum. These organisms have been found in northern and central Mexico (Anaya-López, 2011). However, in the case of C. pubescens, even when the three associated organisms have been found, the presence of P. capsici is associated with intermediate and high cropping intensity in the production zone of the State of Mexico (Table 1).
Molecular identification of the associated organisms. The PCR-amplified DNA fragments of the three associated organisms confirmed the identity of the pathogen, which had been previously morphologically established. The amplified fragment of F. oxysporum had a molecular weight of 900 base pairs (bp), and P. capsici and R. solani had an approximate weight of 650 bp (Figure 2).
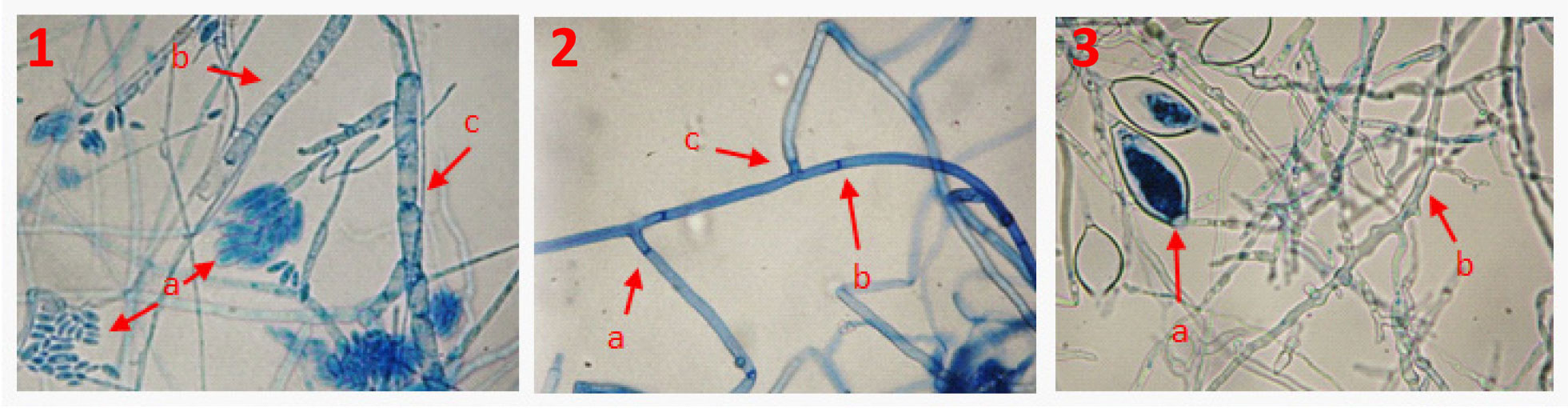
Figure 1 Associated organisms that caused wilt in C. pubescens. 1. F. oxysporum a) microconidia, b) hyaline mycelium, c) septate mycelium. 2. R. solani a) septa close to the point of origin to the hyphal ramification, b) thick and septate mycelium, c) 90° angle. 3. P. capsici a) sporangia, b) hyaline and torulose mycelium.
The sequenced fragments of the F. oxysporum strain were aligned with the F. oxysporum f.sp. lycopersici sequence (accession CM000593.1), and the consensus showed 88% identity with fragments from the region 248-270. The sequenced fragments of the P. capsici strain were aligned with the P. capsici_ LT1534 sequence version 11 accession PcapLT1534_SC064, and the consensus showed 83% identity with fragments from the region 146-259. Similarly, the DNA sequenced fragments of the R. solani strain were aligned with sequences from the NCBI’s gene bank, and its highest level of identity (90%) coincided with that of R. solani, accession number JATN01000256.1 (Version JATN01000256.1 GI: 576995022) with fragments from the region 144-292.
Pathogenicity in vitro. The symptoms observed were necrotic root apex, hypocotyl and/or cotyledons. The F. oxysporum, P. capsici and R. solani isolates were pathogenic to manzano pepper seedlings but symptoms appeared at different time. The expression of symptoms when F. oxysporum was inoculated varied 8-12 days after being cultivated depending on the strain. In the case of seedlings inoculated with P. capsici and R. solani, symptoms appeared 10 days after inoculation.

Figure 2 Electrophoresis in 1.2% agarose gel in TBE for ITS DNA amplification of fragments of F. oxysporum (F), P. capsici (P) and R. solani (R); M lanes correspond to the 100 bp molecular marker.
Statistical differences in incidence and severity were observed on F. oxysporum (P≤ 0.01 colonies, with maximum and minimum values between 34.4 and 92.8% incidence, and 48.8 and 81.6% severity, respectively, and on P. capsici colonies (P≤ 0.05, with maximum and minimum values between 49.2 and 63.6% incidence, and 69.3 and 90.9% severity, respectively, that were associated with the collection site, a fact that may indicate intraspecific heterogeneity associated, among other factors, with pathogens natural spread, such as that reported for powdery mildew (Leveillula taurica) in tomato crops (Solanum lycopersicum) (Guzmán-Plazola et al., 2011), and resistance development by the pathogens due to the selection pressure caused by fungicides (Silva-Rojas et al., 2009). The pathogens incidence was 65% in F. oxysporum, 56. 4% in P. capsici, and 76% in R. solani. On the other hand, the severity values were 81.4% in P. capsici, 64.6% in F. oxyspoum, and 65. 5% in R. solani. These values indicate a more devastating effect caused by oomycetes, which come from a major manzano pepper production zone, where chemical products are used to control diseases, a fact that, according to Silva-Rojas et al. (2009), causes P. capsici to become resistant.
Values higher than 1 in the severity/incidence quotient (Figure 3) indicate a higher level of pathogenicity given that although the pathogens showed a relatively similar incidence, the infection process was faster (severity) on the tissue area. The severity/incidence quotient of F. oxysporum was lower than 1.0 in five out of nine sites where F. oxysporum was found (Figure 3), which represented higher levels of incidence than those of severity, but the average value of the quotient of the nine pathogen colonies was very close to one, which suggests a proportional relation between the pathogen’s incidence and severity. In contrast, the severity/incidence quotient of P. capsici had values higher than 1.0 in all the evaluated colonies (Figure 3), which indicates that the rate of damage was faster even though the incidence was lower. In the case of R. solani, the quotient value was similar to that of F. oxysporum.
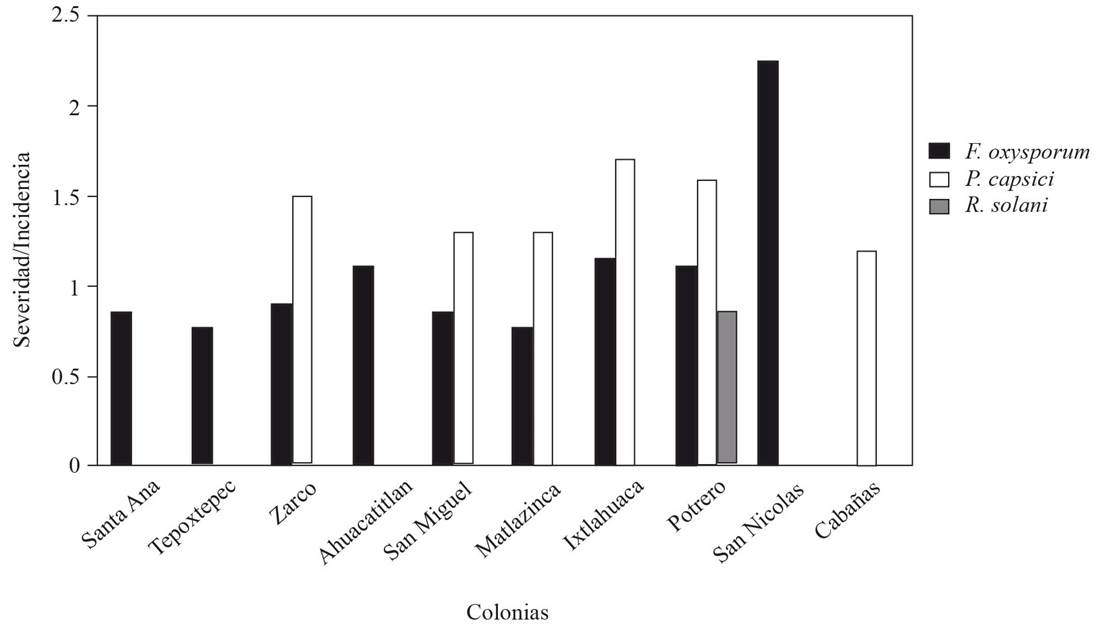
Figure 3 Severity/incidence quotient of F. oxysporum, P. capsici and R. solani in manzano pepper seedlings in 10 production sites of the southern zone of the State of Mexico.
Interaction in multiple infections. The infectious process after inoculations, alone and combined, was faster and symptoms appeared three days after inoculation (dai) (Figure 4). At five dai, plants infected with P. capsici died (Figure 5). Similar results of Capsicum spp. indicate that this is the most lethal pathogen around the world (Lamour et al., 2012). Additionally, reports by Sanzón et al. (2012) describe root necrosis 24 h after inoculation and plant death at day five, as well as evidence of plants being invaded by mycelium and sporangia. However, when P. capsici was combined with other pathogen(s), although no significant differences were found in some treatments (Figure 5), the level of damage was low, which could be due to pathogens competition for nutrients (Abdullah et al., 2017). The average severity values from three-day measurements (Figure 5) showed a significant variation (p≤0.05) in the infection process rate between treatments, which also indicated that the P. capsici inoculum alone was the most severe because of the faster damages accumulation.
In the case of F. oxysporum, seedlings developed brown spots on the hypocotyl five ddi, which, according to Sanzón et al. (2012), are caused by a higher accumulation of polyphenols in the stem tissue that causes the cells in contact to die. Cell wall degradation could be due to activity of lytic enzymes that are produced by the pathogens (Feng et al., 2010).

Figure 4 Damage caused in C. pubescens five dai by: A) F. oxysporum, B) P. capsici, C) R. solani, D) P. capsici + R. solani, E) F. oxysporum + R. solani, F) P. capsici + F. oxysporum and G) F. oxysporum+ P. capsici + R. solani.

Figure 5 Accumulated and average severity of associated organisms alone and combined in C. pubescens plants, 5 days after inoculation. P= P. capsici, P+R= P. capsici + R. solani, F+P= F. oxysporum + P. capsici, F+P+R= F. oxysporum + P. capsici + R. solani, F= F. oxysporum and R= R. solani. The error bars correspond to the standard error. Columns with the same letter are not statistically different (p≤0.05).
When F. oxysporum + P. capsici + R. solani were combined, a co-infection interaction occurred where the pathogenicity was lower compared to that of the infection produced by P. capsici alone (Figure 5). In this interaction complex, the response elicited by one pathogen may be modified in the presence of another pathogen. This complex interaction tends to alter the course of the disease because the co-existing pathogens compete for growth and nutrients in the same host, and suboptimal nutrition leads to a competition whereby some species may dominate (Abdullah et al., 2017). However, the severity and type of competition are determined by nutrient consumption over time (Chesson, 2000).
In the interaction F. oxyporum + R. solani, the pathogenicity was similar to that of F. oxysporum alone, and slightly higher to that of R. solani (Figure 5), which in general terms suggests that when both pathogens are combined, the presence of one of them does not alter the presence of the other. To this regard, Alizon et al. (2013), pointed out that, when the pathogens are combined, there is a wide range of responses from a determined host that have to do with evolutive and ecological aspects from both parts.
Pathogens-genotype interaction. The symptoms in plants inoculated with pathogens, alone and combined, appeared 15 dai. Plants infected with P. capsici showed looseness on the aerial part, necrotic lesions on the internal and external part of the stem crown, and necrotic radicle. Seedlings infected with R. solani showed chlorosis on the aerial part and necrotic roots; when the damage was severe, there was radicle strangulation; González (2008) mentioned non-compact rot with epidermis detachment. Seedlings inoculated with F. oxysporum showed chlorosis and, sometimes, curly foliage and necrotic radicle. Seedlings inoculated with F. oxysporum + P.capsici showed foliar chlorosis and wilt, as well as necrotic stem and radicle.
Seedlings inoculated with F. oxysporum + P.capsici + R. solani showed chlorotic leaves and stem crown, and necrotic radicle. According to Velásquez-Valle et al. (2001), the earliest evident symptom was a slight foliar looseness that was more visible as the day went on, as well as a slight yellowing of foliage. These symptoms were associated with root rot, softening of roots tip, root lesions that varied in size and color (from reddish to brown and black) and short and rotten root (Hamon et al., 2011).
In infections caused by F. oxysporum, there were highly significant differences (P≤0.01) between genotypes with 0-70% variation between the lowest and the highest value, respectively, where the M8 genotype showed resistance, and the M9 genotype showed tolerance to the pathogen (Table 2). In seedlings inoculated with R. solani there were highly significant differences (P≤0.01), with minimum values of 0 % for M8 and maximum values of 76 % for M15. When inoculated with P. capsica, significant differences (P≤0.05) were observed, with 81-95 % variation in susceptibility between M8 and M7, respectively. In pathogen combinations, the intervals between maximum and minimum values were lower. In combinations of F. oxysporum + P.capsici with significant differences (P≤0.05), the maximum and minimum values were 67 % for M8 and 93% for M2; and the combination F. oxysporum + P.capsici + R. solani with highly significant differences (P≤0.01), the minimum and maximum values were 41% for M8 and 95% for nine genotypes (Table 2). The M8 genotype had the lowest level of wilt susceptibility and also showed other distinctive phenotypic differences compared to the rest of the genotypes analyzed (non-reported data), such as dense ramification and small fruits that are characteristic of rustic materials.
Table 2 Susceptibility to wilt damage in 16 manzano pepper genotypes.
| Genotipos | Susceptibilidadx | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | P | R | F*P | F*P*R | ||||||
| M8 | 0 | a | 81 | a | 0 | a | 67 | a | 41 | a |
| M9 | 29 | b | 91 | b | 38 | b | 90 | b | 81 | bc |
| M16 | 48 | c | 95 | b | 48 | bc | 90 | b | 95 | c |
| M15 | 48 | c | 95 | b | 76 | e | 90 | b | 95 | c |
| M11 | 48 | c | 93 | b | 48 | bc | 91 | b | 95 | c |
| M12 | 48 | c | 95 | b | 67 | cde | 85 | b | 83 | bc |
| M10 | 48 | c | 95 | b | 48 | bc | 90 | b | 95 | c |
| M3 | 48 | c | 93 | b | 48 | bc | 90 | b | 95 | c |
| M4 | 48 | c | 95 | b | 48 | bc | 90 | b | 75 | b |
| M5 | 48 | c | 95 | b | 48 | bcde | 90 | b | 93 | c |
| M6 | 48 | c | 95 | b | 73 | e | 85 | b | 90 | bc |
| M7 | 48 | c | 95 | b | 52 | bcd | 90 | b | 95 | c |
| M13 | 57 | c | 95 | b | 61 | cde | 90 | b | 95 | c |
| M2 | 57 | c | 95 | b | 69 | de | 93 | b | 95 | c |
| M14 | 70 | d | 95 | b | 70 | de | 90 | b | 95 | c |
| M1 | 70 | d | 93 | b | 66 | cde | 90 | b | 95 | c |
| p-valorz | 0.0001 | 0.0329 | 0.0001 | 0.0300 | 0.001 | |||||
xF= Fusarium oxysporum; P= Phytophthora capsici; R= Rhizoctonia solani; F*P= Fusarium oxysporum+ Phytophthora capsici; F*P*R= Fusarium oxysporum+ Phytophthora capsici+ Rhizoctonia solani. Medias con letras distintas son significativamente diferentes, de acuerdo a la prueba de Duncan (P≤0.05). zProbabilidad estadística / xF= Fusarium oxysporum; P= Phytophthora capsici; R= Rhizoctonia solani; F*P= Fusarium oxysporum+ Phytophthora capsici; F*P*R= Fusarium oxysporum+ Phytophthora capsici+ Rhizoctonia solani. Medians with different letters are significantly different, according to Duncan’s test P≤0.05). zStatistical probability.
The M8 genotype, and likely, the M9 genotype, were both the least susceptible to wilt damage and can be a potential alternative for introducing the observed resistance in this study to commercial genotypes, such as M3, the most cultivated in the region. Moreover, the differential response of F. oxysporum, P. capscici and R. solani to some of the C. pubescens genotypes analyzed suggests the existence of different resistance mechanisms (Anaya-López, 2011).
Conclusions
Three organisms were detected that cause manzano pepper wilt in the southern zone of the State of Mexico, from which Fusarium oxysporum was the most frequently present, followed by Phytophthora capsici and Rhizoctonia solani.
The molecular identification showed 88% identity of the amplified fragments of F. oxysporum using F. oxysporum f.sp. lycopersici race 2. P. capsici showed 83% identity with P. capsici_ LT1534, version 11, and R. solani had 90% identity with the anastomic group 3 (AG-3).
From the three pathogens identified, Phytophthora capsici causes the highest level of wilt severity in the southern region of the State of Mexico, although Fusarium oxysporum and Rhizoctonia solani also cause plant damage.
The M8 genotype was resistant to F. oxysporum and R. solani, and the M9 genotype could be tolerant to F. oxysporum. The M8 genotype was tolerant to P. capsici and to inoculation with the three pathogens, alone and combined, so it could be a potential genotype for studies about tolerance genes that could be used in genetic improvement programs.
Acknowledgments
The authors wish to thank the Consejo Nacional de Ciencia y Tecnología de México (CONACYT) for the scholarship granted to Alma Janeth Vallejo Gutiérrez for her master’s studies.
REFERENCES
Abawi SG and Pastor-Corrales CMA. 1990. Root rots of beans in Latin America and Africa: Diagnosis, research methodologies, and management strategies. Centro Internacional de Agricultura Tropical (CIAT), Cali, Colombia. 114p. [ Links ]
Abdullah AS, Moffat CS, Lopez-Ruiz FJ, Gibber MR, Hamblin J and Zerihun A. 2017. Host-Multi-Pathogen warfare: Pathogen interactions in co-infected plants. Frontiers in Plant Science 8:1806. https://doi.org/10.3389/fpls.2017.01806 [ Links ]
Alizon S, de Roode JC and Michalakis Y. 2013. Multiple infections and the evolution of virulence. Ecology Letters 16:556-67. https://doi.org/10.1111/ele.12076 [ Links ]
Anaya-López JL, González-Chavira MM, Pineda-Villordo E, Rodríguez-Guerra R, Rodríguez-Martínez R, Guevara-González RG, Guevara-Olvera L, Montero-Tavera V y Torres-Pacheco I. 2011. Selección de genotipos de chiles resistentes al complejo patogénico de la marchitez. Revista Mexicana de Ciencias Agrícolas 3:373-383. Disponible en línea: http://www.scielo.org.mx/pdf/remexca/v2n3/v2n3a6.pdf [ Links ]
Apodaca-Sánchez MA, Zavaleta-Mejía E, García-Espinosa R, Osada-Kawasoe S y Valenzuela-Ureta JG. 2001. Comparación de técnicas para evaluar in vitro la patogenicidad de Fusarium oxysporum f. sp. radicis-lycopersici y efecto de la temperatura. Revista Mexicana de Fitopatología 19:197-2002. Disponible en línea: http://www.redalyc.org/pdf/612/61219210.pdf [ Links ]
Arias AB, Mejía CJ, Estrada MI, Arriaga RM y García VLM. 2017. Caracterización morfológica de híbridos de chile manzano. Revista Mexicana de Ciencias Agrícolas 8:825-836. Disponible en línea: http://www.redalyc.org/html/2631/263152088006/ [ Links ]
Booth C. 1971. The genus Fusarium. Commonwealt Mycological Institute. Kew, Surrey, England. 237 p. [ Links ]
Carrizo GC, Barfuss HJM, Sehr EM, Barboza GE, Rosabelle S, Eduardo A and Ehrenderfer F. 2016. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118:35-51. Disponible en línea: https://www.ncbi.nlm.nih.gov/pubmed/27245634 [ Links ]
Chesson P. 2000. Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics 31:343-366. Disponible en línea: https://doi.org/10.1146/annurev.ecolsys.31.1.343 [ Links ]
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW. InfoStat versión 2016. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. URL. http://www.infostat.com.ar [ Links ]
Erwin DC and Ribeiro OK. 1996. Phytophthora diseases worldwide. The Journal of Agricultural Science 131(2):245-249. https://doi.org/10.1017/S0021859698215796 [ Links ]
Feng B, Li P, Wang H and Zhang X. 2010. Functional analysis of pcpme6 from oomycete plant pathogen Phytophthora capsici. Microbial Pathogenesis 49:23-31. https://doi.org/10.1016/j.micpath.2010.03.004 [ Links ]
Fernández ER, Trapero A y Domínguez J. 2010. Experimentación en agricultura. Junta de Andalucía, Consejería de Agricultura y Pesca. Sevilla, España. 350 p. [ Links ]
Gallegly ME and Honh Ch. 2008. Phytophthora capsici. In identifying species by morphology and ADN fingerprints. APS PRESS. St. Paul, Minnesota, Estados Unidos de América. 165 p. [ Links ]
González GM. 2008. Reseña de “Aspectos de sistemática y biología del complejo Rhizoctonia”. Fitosanidad 12:147-159. Disponible en línea: http://www.redalyc.org/pdf/2091/209115572003.pdf [ Links ]
González-Pérez S, Garces-Claver A, Mallor C, Sáenz de Miera LE, Fayos O, Pomar F, Merino F and Silvar C. 2014. New insights into Capsicum spp. relatedness and the diversification process of Capsicum annuum in Spain. Plos one 9:e116276. Disponible en línea: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4278865/pdf/pone.0116276.pdf [ Links ]
Guigón-López C y González-González PA. 2001. Estudio regional de la enfermedades del chile (Capsicum annuum L.) y su comportamiento temporal en el sur de Chihuahua, México. Revista Mexicana de Fitopatología. 19:49-56. Disponible en línea: http://www.redalyc.org/pdf/612/61219107.pdf [ Links ]
Guzmán-Plazola RA, Fajardo-Franco ML, García-Espinosa R and Cadena-Hinojosa MA. 2011. Desarrollo epidémico de la cenicilla y rendimiento de tres cultivares de tomate en la comarca lagunera, Coahuila, México. Agrociencia 45:363-378. Disponible en línea: http://www.scielo.org.mx/pdf/agro/v45n3/v45n3a9.pdf [ Links ]
Hamon C, Baranger A, Coyne CJ, Mcgee RJ, Le Goff IL, L’anthoëne V, Esnault R and Pilet-Nayel ML. 2011. New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in multiple field and controlled environments from France and the United States. Theoretical and Applied Genetics 123:261-281. DOI: 10.1007/s00122-011-1582-z [ Links ]
Herrera I y Laurentin H. 2012. Evaluación de la esporulación de Fusarium oxysporum f. sp. sesami en dos medios de cultivo y dos metodologías de inoculación en ajonjolí (Sesamum indicum). Revista Científica UDO Agrícola 12:639-643. Disponible en línea: http://www.bioline.org.br/pdf?cg12072 [ Links ]
Kannwischer ME and Mitchell DJ. 1978. The Influence of a fungicide on the epidemiology of black shank of tobacco. Ecology and Epidemiology 68: 1760-1765. Disponible en línea en: https://www.apsnet.org/publications/phytopathology/backissues/Documents/1978Articles/Phyto68n12_1760.pdf [ Links ]
Kousik ChS, Donahoo RS and Hassell R. 2012. Resistance in watermelon rootstocks to crown rot caused by Phytophthora capsici. Crop Protection. 39: 18-25. Disponible en línea: https://www.sciencedirect.com/science/article/pii/S0261219412000907 [ Links ]
Lamour HK, Stam R, Jupe J and Huitema E. 2012. The oomycete broad-host-range pathogen Phytophthora capsici. Molecular Plant Pathology 13:329-337. https://doi.org/10.1111/j.1364-3703.2011.00754.x [ Links ]
Leonian LH. 1922. Stem and fruit blight of peppers caused by Phytophthora capsici. Phytopathology 12:401-408. Disponible en línea: https://www.apsnet.org/publications/phytopathology/backissues/Documents/1972Articles/Phyto62n01_20.PDF [ Links ]
Leslie JF and Sumerell BA. 2006. The Fusarium Laboratory Manual. First edition. State Avenue, Ames, Iowa, USA. Blackwell Publishing. 388 p. Disponible en línea: https://www.wiley.com/en-us/The+Fusarium+Laboratory+Manual-p-9780813819198 [ Links ]
López AGF. 1984. Manejo de Hongos Fitopatógenos. Departamento de Enseñanza e Investigación en parasitología Agrícola. Chapingo, México. pp. 106-115. [ Links ]
Lozano AN, Guzmán-Plazola RA, Zavaleta ME, Aguilar RVH y Ayala EV. 2015. Etiología y evaluación de alternativas de control de marchitez del chile de árbol (Capsicum annuum L.) en la Vega de Metzitlán, Hidalgo, México. Revista Mexicana de Fitopatología 33:31-53. Disponible en línea: http://www.redalyc.org/pdf/612/61240687003.pdf [ Links ]
Martínez GE, Albarracin N, Arcia A, Subero L y Albarracin M. 1996. Pudrición basal del ajo causado por Fusarium oxysporum. Agronomía Tropical 46:265-273. http://hdl.handle.net/20.500.11799/40647 [ Links ]
Martínez EI. 2016. Caracterización morfológica y molecular de 15 colectas de chile manzano (Capsicum pubescens R. y P.) de la región sur del Estado de México. Tesis de Maestría. Centro Universitario UAEM Tenancingo, México. P.43-56. Disponible en línea: http://hdl.handle.net/20.500.11799/40647 [ Links ]
Nelson PE, Tousson TA and Marasa WFO. 1983. Fusarium speies: an illistrated manual for identificaction. The Pennsylvania State University. Pennsylvania, USA. 193 p. [ Links ]
Nora S, Albaladejo RG, González MSC, Robledo-Arnuncio JJ y Aparicio A. 2011. Movimiento de genes (polen y semillas) en poblaciones fragmentadas de plantas. Ecosistemas 20:35-45. Disponible en línea: http://www.redalyc.org/articulo.oa?id=54022121004 [ Links ]
Pickersgill B. 2007. Domestication of plants in the Americas: Insights from Mendelian and molecular genetics. Annals of Botany 100: 925-940. http://doi:10.1093/aob/mcm193 [ Links ]
Ruíz de GJI, Prohens J y Tierno R .2016. Las variedades locales en la mejora genética de plantas. Vitoria-Gasteiz: Servicio Central de Publicaciones del Gobierno Vasco. País Vasco, España. 480 p. [ Links ]
Sanzón GD, Valdovinos PG, Rojas MRI y Zavaleta ME. 2012. Cambios morfológicos en células de chile CM334 inoculado con Phytophthora capsici y con Fusarium oxysporum. Revista Mexicana de Fitopatología 30:66-71. Disponible en línea: http://www.scielo.org.mx/pdf/rmfi/v30n1/v30n1a6.pdf [ Links ]
Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. 2017. Producción nacional de chile alcanza 2.3 millones de toneladas. https://www.gob.mx/sagarpa/prensa/produccion-nacional-de-chile-alcanza-2-3-millones-de-toneladas (consulta, junio 2018). [ Links ]
Segura LS, Zavala RD, Equihua CC, Andrés AJ y Yepez TE. 2009. Los recursos genéticos de frutales en Michoacán. Revista Chapingo Serie Horticultura 15(3): 297-305. Disponible en línea: http://www.scielo.org.mx/pdf/rcsh/v15n3/v15n3a11.pdf [ Links ]
Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación. 2018. Relación de huertos de chile manzano registrados para exportación del Estado de México a los Estados Unidos de América con tratamiento de irradiación. https://www.gob.mx/cms/uploads/attachment/file/379049/CHILE_MANZANO_EDO_MEX_08_23__2018.pdf (consulta, septiembre 2018). [ Links ]
Schubert S, Neubert A, Schierholt A, Sümer A and Zörb C. 2009. Development of salt-resistant maize hybrids: The combination of physiological strategies using conventional breeding methods. Plant Science. 177(3): 196-202. https://doi.org/10.1016/j.plantsci.2009.05.011 [ Links ]
Silva-Rojas HV, Fernandéz-Pavia, SP, Góngora-Canul C, Macías-López BC y Ávila-Quezada GD. 2009. Distribución espacio temporal de la marchitez del chile (Capsicum Annuum L.) en Chihuahua e identificación del agente Causal Phytophthora capsici Leo. Revista Mexicana de Fitopatología. 27: 134-147. Disponible en línea: http://www.redalyc.org/articulo.oa?id=61212195006 [ Links ]
Singlenton L, Mihail JD and Rush MCh. 1992. Methods for research on soil borne Phytopathogenic Fungi. APS Press. St. Paul, Minesota, USA. 264 p. [ Links ]
Uc-Arguelles AK, Pérez-Moreno J, Ayala-Escobar V and Zavaleta-Mejía E. 2017. Antagonism of Saccharicola sp. against phytopathogens of the root of jalapeno pepper (Capsicum annuum). Revista Mexicana de Fitopatología 35:263-283. Disponible en línea: http://www.scielo.org.mx/pdf/rmfi/v35n2/2007-8080-rmfi-35-02-00263.pdf [ Links ]
United States Department of Agriculture Agricultural Research Service (USDA-ARS). 2011. Grin species records of Capsicum. Beltsville, Maryland: National Germplasm Resources Laboratory. https://www.ars.usda.gov/northeast-area/beltsville-md-barc/beltsville-agricultural-research-center/national-germplasm-resources-laboratory/ (consulta, febrero 2018) [ Links ]
Vásquez LA, Tlapa BB, Yáñez MMJ, Pérez PR y Quintos EM. 2009. Etiología de la marchitez del ‘chile de agua’ (Capsicum annuum L.) en Oaxaca, México. Revista Fitotecnia Mexicana 32 (2): 127 - 134. Disponible en línea: http://www.redalyc.org/articulo.oa?id=61011222007 [ Links ]
Velásquez-Valle R, Medina-Aguilar MM y Luna-Ruiz JJ. 2001. Sintomatología y géneros de patógenos asociados con las pudriciones de la raíz del chile (Capsicum annuum L.) en el Norte-Centro de México. Revista Mexicana de Fitopatología 19:175-181. Disponible en línea: http://www.redalyc.org/articulo.oa?id=61219207 [ Links ]
Watanabe T. 2002. Pictorial atlas of soil and seed fungi. Morphogies of cultures fungi and key to species. Second edition. CRC Press. New York Washington, D.C. 500 p. Disponible en línea: http://www.eagriculture.biz/download/Soil/Pictorial%20Atlas%20of%20Soil%20and%20Seed%20Fungi%20%20Morphologies%20of%20Cultured%20Fungi%20and%20Key%20to%20Species.pdf [ Links ]
White TJ, Bruns T, Lee S and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. PCR Protocols. Academic Press. San Diego, CA, USA. P 315-322. Disponible en línea: https://nature.berkeley.edu/brunslab/papers/white1990.pdf [ Links ]
Zhang YL, Jia QL, Li DW, Wang JE, Yin YX and Gong H. 2013. Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici Leonian. International Journal of Molecular Sciences 14: 8985-9004. https://doi.org/10.3390/ijms14058985 [ Links ]
Received: September 27, 2018; Accepted: November 19, 2018











 text in
text in 


