Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.36 no.2 Texcoco may./ago. 2018
https://doi.org/10.18781/r.mex.fit.1712-2
Phytopathological notes
Anatomic alterations and hyperplasia induced by Euphorbia mosaic virus Yucatan Peninsula isolate at the meshophyll
1Unidad de Bioquímica y Biología Molecular de Plantas, Centro de Investigación Científica de Yucatán, Calle 43 No. 130, Colonia Chuburná de Hidalgo CP. 97200. Mérida, Yucatán. México.
Euphorbia mosaic virus isolated from the Yucatan Peninsula is a bipartite begomovirus with a wide range of experimental hosts, including species of economic importance, such as bean, chilli and tomato, among others. This virus produced severe symptoms on Nicotiana benthamiana leaves, leaf blade deformations, loss of tissue and cell identity, as well as changes in the mesophyll structure. Unlike other bipartite begomoviruses, EuMV-YP induced cell proliferation at the mesophyll; it also positively regulated the expression of cycA1, cycA2 and cdkB2.2, genes involved in the G2/M transition of the mitotic cell cycle, and localized at the mesophyll. This is the first example of a bipartite begomovirus capable of inducing cell proliferation.
Key words: geminivirus; cell cycle
Euphorbia mosaic virus aislamiento Yucatan Peninsula es un begomovirus bipartita con una amplia gama de hospedantes experimentales, que incluyen especies de importancia económica, como frijol, chile y tomate, entre otras. Este virus provocó síntomas severos en hojas de Nicotiana benthamiana, pérdida de identidad celular y tisular, deformaciones de la lámina foliar y de la nervadura central, cambios en la estructura del mesófilo y proliferación celular, a diferencia de otros begomovirus bipartitas. Activó la expresión de los genes cycA1, cycA2 y cdkB2.2, implicados en la transición G2/M del ciclo celular mitótico y se localizó en el mesófilo de las hojas. EuMV-YP es el primer ejemplo de un begomovirus bipartita capaz de inducir la proliferación celular.
Palabras clave: geminivirus; ciclo celular
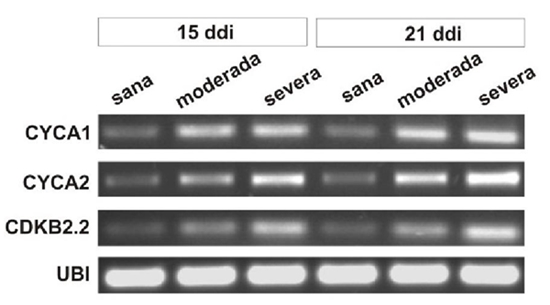
Euphorbia mosaic virus, isolated from the Yucatan Peninsula (EuMV-YP) is a bipartite begomovirus with a wide range of experimental hosts. The gene silencing induced by EuMV-YP in Nicotiana benthamiana begins in the-midribs and extends to the rest of the leaf (Villanueva et al., 2013), which indicates that it is capable of reaching the mesophyll. Most bipartite begomoviruses have specific hosts in which they are restricted to the phloem. The Tomato golden mosaic virus (TGMV) and the African cassava mosaic virus (ACMV) can invade the mesophyll. There is a correlation in its capability to exit the phloem and to produce severe symptoms in N. benthamiana (Wege et al., 2001). The replication of geminiviruses takes place in the nuclei of differentiated cells, and for this, they induce their entry into the cell cycle. The bipartite begomoviruses and mastreviruses promote DNA synthesis without undergoing cell division (revised by Hanley et al., 2013); whereas curtoviruses induce mitosis, producing enations in the leaves midribs (Park et al., 2004; Latham et al., 1997). This investigation posed the question of whether EuMV-YP is able to reach cells of the N. benthamiana mesophyll and if its tissue tropism correlates with the severity of the symptoms produced. To inoculate with the wild virus, previously constructed infective clones were used (Villanueva et al., 2013), and to locate the virus, expression vector pEuMV-YPDAV1:GFP was constructed. The combinations of plasmids used per treatment are indicated in Table 1. In each treatment, 20 N. benthamiana plants with four true leaves were inoculated, as reported previously (Villanueva et al., 2013); in this investigation, a pressure of 70 psi was applied at a distance of 6 cm, as well as 1 μm gold particles covered with 5 μg of each plasmid. Fifteen days post inoculation (dpi), leaves number nine and 10 were harvested . They are were named “systemic leaves,”in this manuscript, since they emerged after inoculation Paraffin inclusions of fixed tissues, and 8-10 μm transversal cuts of the center of the leaves were made, as reported previously (Moo et al., 2012). Images were taken using a 40x lens of an Olympus FV100 SW laser confocal microscope, and they were processed using the FV10 ASW software version 4.1. The GFP fluorescence was captured with 488/507 nm wavelengths of excitation/emission, respectively. For the histological analysis, the sections were dyed with toluidine blue. Images were taken of transversal sections and of the midsections of leaves with systemic infections using the 6.5x lens of a Nikon E200 microscope. The leaf’s thickness was measured and the number of cells per mm2 was counted in four plants with moderate to severe symptoms at 15dpi. The number of cells and the thickness of the leaf blade were graphed using 12 individual values. We tested the null hypothesis that the means of plants with severe and moderate symptoms are equal to healthy plants. F tests were performed on two samples to determine the homogeneity of variances. The statistical significance between the pairs of means (severe or moderate symptoms vs healthy plants) was obtained using the Student’s t-test on two samples, assuming unequal variances, using Excel’s Data Analysis ToolPak program. For the gene expression studies, leaves with systemic infections were taken from three plants, frozen, and ground together in liquid nitrogen. Total RNA was extracted with the RNeasy Plant mini kit (Qiagen) and a treatment with DNase I. The first cDNA strand was synthesized with 500 ng of total RNA, Oligo-dT [d (T) 23VN] and the ProtoScript M-MuLV kit, following the manufacturer protocol (New England Biolabs, U.S.A.), and the final volume was adjusted to 50 μL. For the second strand,0.2 μM of specific primers were used for genes cycA1, cycA2, cdkB2.2 (Caracuel et al., 2012) and ubi3 (Kotakis et al., 2010), 5 μL of the cDNA, 0.2 mM dNTPs, 1.25 units of Taq polymerase and Thermo Pol buffer (New England Biolabs) in 25 μL, with the program: 94 °C for 5 min / 28 cycles at 94 °C for 30 s, 56 °C for 60 s and 72 °C for 1 min / 72 °C for 10 min. We separated 10 μL in an agarose gel at 0.8% stained with ethidium bromide. GFP was found directly in all types of mesophyll cells, such as the upper epidermis and the parenchyma in palisade (Figure 1A), the spongy parenchyma and the lower epidermis (Figure 1C) of the leaves with systemic infections 15 dpi. No GFP was found in healthy plants (Figure 1B and 1C). GFP fluorescence was low and was not directly detectable in the phloem of the midrib and in other cells of the vascular bundles, although it was found using immunodetection (data not shown). These results show that EuMV-YP is capable of invading all types of mesophyll cells in N. benthamiana in a high proportion, just as ACMV and TGMV (Wage et al., 2001). EuMV-YP also caused severe symptoms in N. benthamiana; at 15 dpi, plants were found with different degrees of severity: seven mild, eight moderate and five severe. The leaves with systemic infections from plants with severe symptoms presented strong downward curling of the margins, as well as abundant crinkling or bulging (Figure 2A, 2B and 2C); these symptoms remained until 21 dpi. At 25 dpi, chlorotic mosaics appeared and at 28 dpi, the symptoms became attenuated (Figure 2D), and remained mitigated until the end of the experiment. These results show that, like ACMV and TGMV, EuMV-YP is capable of inducing severe symptoms in N. benthamiana. Three genetic determinants have been described that confer the tissue tropism of TGMV (Morra and Petty, 2000). EuMV-YP belongs to a different taxonomic clade (Hernández et al., 2007), and therefore the genetic determinants that give it the capability of invading the mesophyll and of producing severe symptoms can be different. The bulges observed in the leaves of plants with severe symptoms suggested that EuMV-YP caused anatomic alterations in the leaves (Figure 2A), as well as a high frequency of cell division events (Figure 1A). To characterize this finding, a thorough histological analysis was carried out on the leaves in discreet regions throughout the entire leaf blade (Figure 3A). In healthy plants, a typical N. benthamiana tissue organization was observed; upper and lower epidermis and the palisade parenchyma of one single layer of cells, the latter with elongated anticlinal cells; the spongy parenchyma with two or three layers of spherical cells (Figure 4F). In infected plants, areas were noticed with changes in the morphology of cells and thickened regions (Figure 4B, D, E and F), accounting for 10 and 50% of the leaf in plants with moderate and severe symptoms, respectively. The average value of the thickened regions (altered) in plants that displayed severe symptoms was significantly different to that of the regions without changes (normal) in Student’s t tests (t ≤ 7.55, P << 0.01); the same happened in regions vs normal regions of plants with moderate symptoms (t ≤ 2.71, P ≤ 0.02) (Figure 3B Left). The number of cells was significantly higher in plants with severe or moderate symptoms in comparison with those in healthy plants (t ≤ 6.18, P <0.01, t <3.87, P <= 0.01, respectively), with increases of 50 and 100% of the plants with moderate and severe symptoms, respectively (Figure 3B Right). Cell proliferation was observed in the palisade and spongy parenchyma (Figure 4), which commonly had four layers of cells (Figure 4E and 4F). However, in plants with the most severe symptoms, up to 10 layers were found (data not shown). Certain regions of the leaves presented alterations in cell morphology, particularly of the palisade and spongy parenchyma (Figure 4B y 4B). Also, shpere-shaped and small cells were observed in the regions with the greatest cell proliferation (Figure 4F). These results confirm that the bulges in the leaves are a product of the induction of cell division in the N. benthamiana mesophyll. Similar results were found when expressing protein C12 of the Chrysanthemum virus B (CVB) in N. tabacum leaves, which is a determinant of pathogenicity and virulence. CVB causes curling and cell proliferation in the leaves of chrisanthemum, its natural host (Lukhovitskaya et al., 2014). In fact, many viruses produce similar symptoms, though no histological studies have been carried out to reveal the nature of the symptoms, and hence the relevance of this study. It is worth pointing out that EuMV-YP is the first example of a geminivirus that can induce cell division in the palisade and spongy parenchyma, and is therefore an interesting model to study the gene determinants that are inducing this phenomenon. On the other hand, there are several reports on the induction of cell division in the phloem and phloem-associated cells in the leaf midribs. Such is the case of Curtoviruses and the monopartite begomoviruses, amongst the geminiviruses (Park et al., 2004; Srivastava et al., 2017) and RNA viruses (Xie et al., 2014; Shen et al., 2016); in all these cases, the induced cell proliferation leads to the production of tumors in the leafmidribs. In consistency with the previous results, the expression of genes cycA1, cycA2 and cdkB2.2 increased in the leaves of infected plants at 15 and 21 dpi in comparison with those for healthy plants (Figure 5), while gene ubi3 remained unchanged. The positive regulation of genes that participate in the entry into the mitotic phase (M) (Breyne et al., 2002) is consistent with the cell increases in number found in plants with moderate or severe symptoms. Similar results were observed in plants infected with BCVT (Park et al., 2004), CBV (Lukhovitskaya et al., 2014) and the Rice black-streaked dwarf virus (RBSDV;Shen et al., 2016). On the other hand, the bipartite begomoviruses Cabbage leaf curl virus (CaLCuV) and TGMV, activate the entry to the endocycle in Arabidopsis thaliana and in N. benthamiana, respectively (Nagar et al., 2002; Ascencio et al., 2008). CaLCuV activates the expression of most of the genes that participate in the entry to the cell cycle in the phase of DNA synthesis and suppresses the expression of most genes associated to M phase or cell division such as CDKB2 in A. thaliana (Ascencio et al., 2008), which explains CaLCuV inducing endoreduplication and not cell division. EuMV-Jal and EuMV-YP isolates, which produce similar symptoms in several experimental hosts and are incompatible in replication, have acquired unique sequences in their intergenic regions that set them apart from phylogenetically related viruses (Gregorio et al., 2010). EuMV-YP may have lost the sequences needed to block the G2/M checkpoint or it may have acquired gene determinants to induce a complete mitotic cycle, whether by mutation or the incorporation of genomic sequences from other viruses. This is yet to be determined. In conclusion, EuMV-YP is capable of producing severe symptoms in N. benthamiana and of reaching the mesophyll, where it induces mitosis and changes in cell morphology, as well as the expression of genes related to cell division.
Table 1 Plasmids used per treatment
|
Tratamiento/treatment |
Plásmidos/plasmids |
Ensayo/Assay |
|
|
xTT |
ySH |
||
|
Testigo/control |
pBluescript II SK+ |
+ |
+ |
|
Virus silvestre/ Wild type virus |
pEuMV-YP:HA + pEuMV-YP:HB |
+ |
- |
|
Vector GFP / GFP vector |
pEuMV-YPDAV1:GFP |
+ |
- |
|
EuMV-YP GFP |
pEuMV-YPDAV1:GFP + pEuMV-YP:HB |
+ |
- |
|
EuMV-YP + EuMV-YP GFP |
pEuMV-YP:HA + pEuMV-YP:HB + pEuMV-YPDAV1:GFP |
+ |
- |
xTT = Tissue Tropism. ySH=Symptoms and Histology

Figure 1 Tissue tropism of EuMV-YP in N. benthamiana. Plants inoculated with the wild virus + EuMV-YP GFP at 15 ddi (A, C) and healthy plants (B, D). Palisade (A, B) and spongy parenchyma (C, D). Bars: 5 µm

Figure 2 Severe symptoms produced by EuMV-YP. EuMV-YP (A-D) and healthy plants (E-H), at 15 (A-C, E-G) and 28 dpi (D and H). Dorsal (B and F) and ventral (C and G) views.

Figure 3 Quantitative analysis of the effect of EuMV-YP. Cross section of a leaf (A). Graphs of the thickness of the leaf blade and number of cells (B) in plants with moderate or severe symptoms, or healthy plants. Includes standard deviation error bars.
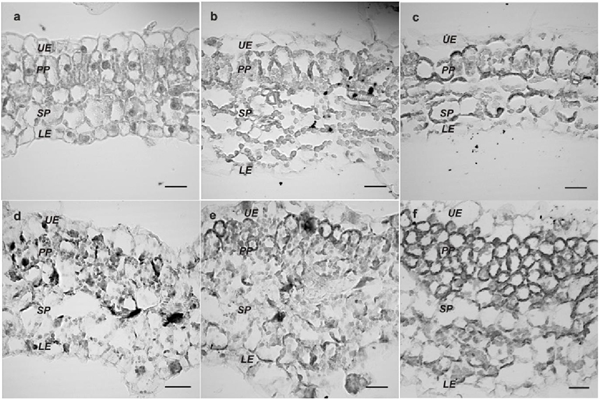
Figure 4 Anatomical changes induced by EuMV-YP. Healthy plants (A) or with severe symptoms (B-F). Changes in cell morphology in spongy (SP) and in palisade parenchyma (PP) (B-C). Upper (UE) and lower (LE) epidermis. 680x scale. Bars: 20 μm.
Acknowledgements
To Oscar Moreno and Hernán Villanueva for the hemidimers and the VIGS vector; to Jorge Ramírez Prado for his support with the statistical analyses. To CONACYT for the funds given to project #98920 to LL0 and for the PhD scholarship #46688 to NGT.
REFERENCES
Ascencio IJT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R and Hanley BL. 2008. Global analysis of arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiology 148:436-454. DOI: 10.1104/pp.108.121038 [ Links ]
Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, and Zabeau M. 2002. Transcriptome analysis during cell division in plants. Proceedings of the National Academy of Sciences 99:14825-14830. https://doi.org/10.1073/pnas.222561199 [ Links ]
Caracuel Z, Lozano DR, Huguet S, Arroyo MM, Rodríguez NEA and Bejarano ER. 2012. C2 from beet curly top virus promotes a cell environment suitable for efficient replication of geminiviruses, providing a novel mechanism of viral synergism. New Phytologist 194:846-858. DOI: 10.1111/j.1469-8137.2012.04080.x [ Links ]
Gregorio JJ, Bernal AA, Bañuelos HB, Alpuche SAG, Hernández ZC, Moreno VO, Frías TG, Argüello AG. 2010. Analysis of a new strain of Euphorbia mosaic virus with distinct replicationspecificity unveils a lineage of begomoviruses with short Rep sequences in the DNA-B intergenic region. Virology Journal 7:275. https://doi.org/10.1186/1743-422X-7-275 [ Links ]
Hanley BL, Bejarano ER, Robertson D and Mansoor S. 2013. Geminiviruses: masters at redirecting and reprogramming plant processes. Nature Reviews Microbiology 11:777-788. DOI: 10.1038/nrmicro3117 [ Links ]
Hernández ZC, Idris AM, Carnevali G, Brown JK and Moreno VO. 2007. Molecular characterization and experimental host range of Euphorbia mosaic virus-Yucatan Peninsula, a begomovirus species in the Squash leaf curl virus clade. Plant Pathology 56:763-770. DOI: 10.1111/j.1365-3059.2007.01652.x [ Links ]
Kotakis C, Vrettos N, Kotsis D, Tsagris M, Kotzabasis K y Kalantidis K. 2010. Light intensity affects RNA silencing of a transgene in Nicotiana benthamiana plants. BMC Plant Biology 10:220. https://doi.org/10.1186/1471-2229-10-220 [ Links ]
Latham JR, Saunders K, Pinner MS and Stanley J. 1997. Induction of plant cell division by beet curly top virus gene C4. The Plant Journal 11:1273-1283. DOI: 10.1046/j.1365-313X.1997.11061273.x [ Links ]
Lukhovitskaya NI, Solovieva AD, Boddeti SK, Thaduri S, Solovyev AG and Savenkov EI. 2013. An RNA virus-encoded zinc-finger protein acts as a plant transcription factor and induces a regulator of cell size and proliferation in two tobacco species. The Plant Cell 25:960-973. DOI: 10.1105/tpc.112.106476 [ Links ]
Morra MR and Petty IT. 2000. Tissue specificity of geminivirus infection is genetically determined. The Plant Cell 12: 2259-2270. https://doi.org/10.1105/tpc.12.11.2259 [ Links ]
Moo LRC, González AK, Rodríguez LC, Suarez V, Castaño E. 2012. Expression of RNA polymerase IV and V in Oryza sativa. Electronic Journal of Biotechnology 15:9-9. DOI: 10.2225/vol15-issue2-fulltext-8 [ Links ]
Nagar S, Hanley BL and Robertson D. 2002. Host DNA replication is induced by geminivirus infection of differentiated plant cells. The Plant Cell 14:2995-3007. http://doi.org/10.1105/tpc.005777 [ Links ]
Park J, Hwang H, Shim H, Im K, Auh CK, Lee S and Davis KR. 2004. Altered cell shapes, hyperplasia, and secondary growth in Arabidopsis caused by beet curly top geminivirus infection. Molecules and Cells 17:117-124. Disponible en línea: https://www.ncbi.nlm.nih.gov/pubmed/15055537 [ Links ]
Shen J, Chen X, Chen J and Sun L. 2016. A phloem-limited fijivirus induces the formation of neoplastic phloem tissues that house virus multiplication in the host plant. Scientific Reports 6:29848. http://doi.org/10.1038/srep29848 [ Links ]
Srivastava A, Agrawal L, Raj R, Jaidi M, Raj SK, Gupta S, Dixit R, Singh PC, Tripathi T, Sidhu OP, Singh BN, Shukla S, Chauhan PS and Kumar S. 2017. Ageratum enation virus Infection Induces Programmed Cell Death and Alters Metabolite Biosynthesis in Papaver somniferum. Frontiers in Plant Science 8:1172. DOI: 10.3389/fpls.2017.01172 [ Links ]
Villanueva AH, Us CR, López 0L, Robertson D, Guerra PO, Minero GY and Moreno VO. 2013. A new virus-induced gene silencing vector based on Euphorbia mosaic virus-Yucatan peninsula for NPR1 silencing in Nicotiana benthamiana and Capsicum annuum var. Anaheim. Biotechnology Letters 35:811-823. https://doi.org/10.1007/s10529-013-1146-1 [ Links ]
Wege C, Saunders, K, Stanley J and Jeske H. 2001. Comparative analysis of tissue tropism of bipartite geminiviruses. Journal of Phytopathology 149:359-368. DOI: 10.1046/j.1439-0434.2001.00640.x [ Links ]
Xie L, Lv MF, Zhang HM, Yang J, Li JM, Chen JP. 2014. Tumours induced by a plant virus are derived from vascular tissue and have multiple intercellular gateways that facilitate virus movement. Journal of Experimental Botany 65:4873-4886. DOI: 10.1093/jxb/eru254 [ Links ]
Received: December 05, 2017; Accepted: February 05, 2018











 texto en
texto en