Services on Demand
Journal
Article
Indicators
Related links
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.36 n.1 Texcoco Jan./Apr. 2018
https://doi.org/10.18781/r.mex.fit.1710-3
Scientific articles
Root endophyte bacteria in drought-tolerant and drought-susceptible maize lines
1 Fitosanidad-Fitopatología, Colegio de Postgraduados. Km. 36.5 Carretera Mexico-Texcoco. Montecillo, CP. 56230, Texcoco, Estado de Mexico.
2 Department of Plant Pathology, School of Environmental Sciences, University of Guelph, 50 Stone Road East, Guelph, Ontario, N1G 2W1, Canada.
3 Laboratorio de Parasitología Agrícola, Universidad Autónoma Chapingo, Km 38.5 Carretera México-Texcoco, Chapingo, CP. 56230, Texcoco, Estado de México.
Maize (Zea mays) ranks second as food in the world and drought limits its productivity. Plants harbor endophytic bacteria that influence health and drought tolerance. The goal of this research was to estimate the density and diversity of cultivable endophyte bacteria from the root system of seven homozygous maize drought-tolerant and seven drought-susceptible lines in three locations of Mexico during three crop cycles. The density and diversity of bacterial populations was assessed by direct counting on plates and identified by PCR. The results identified three groups of endophytic bacteria: 1) highly frequent (Bacillus subtilis, Bacillus megaterium y Pseudomonas geniculata), 2) frequent (Bacillus firmus, Pseudomonas hibiscola y Sinorhizobium meliloti) y 3) low frequency (Acinetobacter soli, Stenotrophomonas maltophila y Burkholderia gladioli. The analysis of variance (ANOVA) showed significant differences (p≤0.05) in density (Log10 CFU g-1 root) of population by location, crop cycle, days after sowing and maize lines. The density of Bacillus subtilis, Pseudomonas hibiscola at El Batán and Bacillus megaterium, Sinorhizobium meliloti in Tlaltizapán, were significantly higher in drought-tolerant maize lines compared to drought-susceptible lines.
Key words: Zea mays; endophytic bacteria; bacterial diversity; 16S rADN
El maíz (Zea mays) ocupa el segundo lugar como alimento en el mundo y la sequía limita su productividad. Las plantas albergan bacterias endófitas que influyen en la sanidad y tolerancia a la sequía. El objetivo de esta investigación fue estimar la densidad y diversidad de las bacterias endófitas cultivables de la raíz en siete líneas homocigóticas de maíces tolerantes y siete susceptibles a sequía en tres localidades de México durante tres ciclos del cultivo. La densidad y diversidad de las poblaciones bacterianas se evaluó mediante conteo directo en placas y se identificaron por PCR. Los resultados identificaron tres grupos de bacterias endófitas: 1) altamente frecuentes (Bacillus subtilis, Bacillus megaterium y Pseudomonas geniculata), 2) frecuentes (Bacillus firmus, Pseudomonas hibiscola y Sinorhizobium meliloti) y 3) baja frecuencia (Acinetobacter soli, Stenotrophomonas maltophila y Burkholderia gladioli. El análisis de varianza (ANOVA) mostró diferencias significativas (p≤0,05) en la densidad (Log10 UFC g-1 de raíz) de población por localidad, ciclo de cultivo, días después de siembra y líneas de maíz. La densidad de Bacillus subtilis, Pseudomonas hibiscola en la localidad de El Batán y Bacillus megaterium, Sinorhizobium meliloti en Tlaltizapán, fueron significativamente mayores en las líneas de maíz tolerantes que en las susceptibles a sequía.
Palabras clave: Zea mays; bacterias endofíticas; diversidad bacteriana; 16S rADN
Maize (Zea mays) is currently the second most important food staple in terms of energy sources, and provides a protein content of approximately 9.2% to human nutrition (FAO, 2016). Mexico is in the seventh place in maize production, with approximately 25 million tons a year (FIRA, 2016). Maize productivity is limited by several biotic stresses (pests and diseases) and abiotic stresses (drought, nutrient deficiency, salinity and high temperatures) (Grover et al., 2007). Drought is the meteorological term for water scarcity, and is one of the major environmental stress factors that affect germination, plant vigor and productivity of agricultural crops (Kamara et al., 2003; Wilkinson and Davies, 2010). A possible alternative for tackling this problem is to develop knowledge about plant microorganisms that play an important role in the expression of abiotic stress resistance (Gond et al., 2015).
Endophytic microorganisms are bacteria, fungi or viruses that live all or part of their lives in plants’ internal tissues without damaging their hosts. They establish a symbiotic interaction that modulates the health of plants, as well as their ability to adapt to different environmental stress factors (Hardoim et al., 2015). Endophytic bacteria are a subgroup of the rhizosphere and rhizoplane bacterial community that colonizes the internal root tissue of the host plant; this gives them ecological advantages over other populations that colonize epiphytically. Diverse species of Gram-positive and Gram-negative endophytic bacteria have been isolated from different types of tissue from many plant species (Rosenblueth and Martínez-Romero, 2006).
There is evidence of the positive effect of endophytic microorganisms through the expression of mechanisms such as antagonism, induced systemic resistance (RSI) and acquired systemic resistance (RSA), plant growth promoters and induced adaptation response to environmental stresses (Liu et al., 2013). The relationship between the host plant and the endophytic bacterial community reflects a co-evolution in the colonization process influenced by the genotype, growth stage, physiological condition and plant tissue, as well as by soil characteristics, agronomic practices and environmental conditions such as temperature, water and nutrient supply (Higgins et al., 2007).
When maize is inoculated with these microorganisms, this increases germination, plant height, root and aboveground mass and improves yield (Morales et al., 2011). It has also been shown that inoculating maize with these bacteria efficiently promotes drought stress tolerance (Fan et al., 2015) as a result of the increase in root length and biomass, which in turn improves water and nutrient absorption (Naseem and Bano, 2014; Yasmin et al., 2013). Endophytic bacteria can induce tolerance to abiotic stresses such as salinity and drought, while some populations confer tolerance to specific stress factors and are responsible for the survival of plants under these particular environmental conditions (Gond et al., 2015; Montañez et al., 2011). Thus, the microorganisms that establish positive interactions with plant roots play a key role in cropping systems and have promising biotechnological potential to be used in sustainable agriculture.
In Mexico there are no studies on endophytic bacterial communities in roots of drought tolerant or drought susceptible maize lines. Such endophytic bacteria could be used to develop future biotechnological strategies such as inducers of drought tolerance and other abiotic and biotic factors that limit crop productivity. Therefore, the objective of this study was to estimate the density and diversity of endophytic bacterial populations in roots of homozygous (S6) drought tolerant and drought susceptible maize lines previously identified as such under field conditions and sown at three Mexican locations during three crop cycles.
Materials and methods
Sampling site. Seven drought-tolerant homozygous maize lines (CLQRQ108, CML384, CML445, CML544, DTMA90, DTMA224, DTMA256) and seven drought-susceptible homozygous maize lines (CML181, DTMA34, DTMA41, DTMA43, DTMA109, DTMA144, DTMA182) were sown, which were selected because they were inbred due to their uniform response to drought. Each line was sown with three replications at the experiment stations of the International Maize and Wheat Improvement Center (CIMMYT) in Tlaltizapán, Morelos (18.68 N; 99.11 O), El Batán, Texcoco, Mexico (19.53 N; 98.85 W), and Agua Fría, Puebla (20.5 N; 97.6 W) during the 2012 summer crop cycle (V-2012), 2012 winter cycle (I-2012) and 2013 autumn cycle (V-2013). Samples of the internal tissue formed in the apical root area of three plants per line were taken for each location and crop cycle 25, 52 and 75 days after sowing (dds) to be analyzed at CIMMYT’s Maize Pathology Laboratory at El Batán, State of Mexico. The size of the sample was determined by the method proposed by Cochran (1982).
Isolation of endophytic root bacteria. Bacterial isolates from the internal tissue of maize roots that had been superficially sterilized were defined as endophytic strains. Symptomless maize roots were washed with sterile distilled water and cut in 2-cm pieces. 10 g of roots were sterilized superficially for 5 min in sterile glass jars and washed with 70% ethanol for 5 min, 0.53% sodium hypochlorite for 10 min and then rinsed three times with sterile distilled water. After the last washing with sterile distilled water, 100 µL from each jar were sown in Petri dishes containing NB culture medium (1L): 15 g bacteriological agar (BIOXON® Mexico), 5 g peptone (BD DIFCOTM USA), 3 g yeast extract (DIBICO® Mexico, NaCl JT BAKER® Mexico) and incubated at 28±1oC from 2 to 5 days. The absence of bacterial growth in the culture medium confirmed that the root surface had been efficiently sterilized. The roots were ground in cold mortars using 20 mL of a sterile buffer solution (50 mM KH2PO4, 150 mM NaCl, pH 7.6). The resulting suspensions were diluted in series (1:9) from 100 to 10-3; then 100 µL of each dilution and three replications were sown in NB medium and incubated at 28 ºC for 24 h under continuous light. The different bacterial morphotypes were counted and classified according to their color, shape, texture and type of growth. The bacterial growth was considered representative of cultivable endophytic bacteria in maize roots. Bacterial strains were preserved by freezing in NB medium and 20% glycerol for subsequent studies.
Population density of endophytic bacteria. The population density of bacteria per root tissue was estimated by directly counting the colonies on the plate. The number of bacterial colonies isolated from the 14 maize lines was counted using a colony counter (Quebec®, Darkfield Colony Counter). The bacterial population of root tissue samples was expressed in terms of colony forming units (UFC g-1 of root tissue). The resulting data were converted to Log10 UFC g-1 of root and an analysis of variance (ANOVA) was performed using a factorial analysis design to separate the media by DMS (α=0,05) using the SAS statistical program (Statistical Analysis System version 9.1.3 SAS Institute, Cary, NC, USA) according to the crop cycle, location, days after sowing (dds) and the studied drought-tolerant or drought-susceptible maize lines.
Amplification of the 16S rDNA gene and identification of endophytic bacteria. Bacterial DNA was extracted using the protocol described by Mahuku (2004) with the following modifications: bacterial cells were obtained by growing them in NB medium at 28 °C for 48 h. After suspending the precipitate in 100 µL of 1X TE, 2 µL of RNAsa were added (1 mg mL-1) and then incubated in a water bath at 37 °C for 1 h. The DNA quality was verified by electrophoresis in agarose gel. Bacteria were identified by partially amplifying the 16S rADN gene using universal primers 27F (5´AGAGTTTGATCMTGGCTCAG-3´) and 1492R (5´TACGGHTACCTTGTTACGACTT-3´) under the PCR conditions described by Galkiewicz and Kellogg (2008). DNA amplification and sequencing were carried out using Macrogen (DNA Sequency Service. Korean Biotechnology Company), and the sequences obtained were aligned with sequences kept at the GenBank of (NCBI) using the BLASTn program (http://www.ncbi.nlm.nih.gov/BLAST/).
Results and discussion
Taxonomic identification of endophytic bacteria. Through partial sequencing of the 16S rADN gene, 22 endophytic bacteria species were identified in roots of both groups of maize lines. The greatest abundance of bacteria was associated with the Proteobacteria phyla followed by Firmicutes and Bacteroidetes. Within Firmicutes, 100% of strains were identified as Bacillus sp., and within the class Gammaproteobacteria, 33% were identified as Pseudomonas sp.
The diversity and richness of bacteria naturally associated with maize roots is wide, and their estimation depends on the analytic method used. In this study, a dependent culture approach was used, and the results coincide with previous studies that found that the Proteobacteria phylum was the most predominant of the endophytic bacterial populations isolated from maize roots, stems and leaves (Rosenblueth and Martínez-Romero, 2006). Other studies of the diversity of endophytic communities in maize roots using dependent and independent culture methods also found that the bacteria most frequently associated with this crop are Firmicutes (Bacillus), Gammaproteobacteria (Pseudomonas) (Pereira et al., 2011) and Burkholderia spp. (Ikeda et al., 2013). By analyzing these populations using gas chromatography and fatty acid profiles, Bacillus pumilus, B. subtilis, Pseudomonas aeruginosa and P. fluorescens were identified as the most predominant species in maize stalks (Rai et al., 2007), but when using pyrosequencing, the Proteobacteria, Bacteroidetes and Actinobacteria phyla were identified as being the most abundant in the maize rhizosphere (Li et al., 2014).
Maize research has shown that these populations could be used as bacterial inoculants for controlling drought stress, and suggests that drought tolerance is induced by the production of phytohormones such as abscisic, gibberellic and indole-3-acetic acids, cytokinins, enzymes such as ACC deaminase, production of bacterial exopolysaccharides and systemic tolerance induction (Dimkpa et al., 2009; Kim et al., 2012; Timmusk et al., 2014; Yang et al., 2009). In other crops, they have been identified as tolerance inductors in wheat (Triticum sativum) and legumes (Vigna radiata) when they were inoculated in seed, for they promoted an increase in the levels of regulation of genes related to drought tolerance and the activity of diverse enzymes (Kasim et al., 2013; Saravanakumar et al., 2011).
Based on the frequency of the isolates, three groups of endophytic bacteria in the roots of drought tolerant and drought susceptible maize stand out in this study: 1) highly frequent (Bacillus subtilis, Bacillus megaterium and Pseudomonas geniculata), isolated during the three crop cycles at the three locations, 2) frequent (Bacillus firmus, Pseudomonas hibiscola and Sinorhizobium meliloti), isolated during the three crop cycles at two locations, and 3) low frequency (Acinetobacter soli, Stenotrophomonas maltophila and Burkholderia gladioli), isolated only at two locations and not in all crop cycles (Table 1).
Table 1 Endophyte bacteria species identified in roots of 14 maize lines in three locations during three crop cycles.
| ID | Endófito | Línea | Identidad de Nucleótidos | Clase | Localidad/ciclo de cultivo | ||
| T/S | TL | AF | EB | ||||
| AF101 | Acinetobacter soli Z | T, S | 96% (KU551890) | γ Proteobacteria | 1 | 1 | |
| AF105 | Agrobacterium tumefaciens | T, S | 97% (KX518841) | α Proteobacteria | 1 | ||
| TL007 | Bacillus asahii | T, S | 95% (KU551893) | Firmicutes | 1, 2, 3 | 1 | |
| TL008 | Bacillus firmus Y | T, S | 98% (KU551896) | Firmicutes | 1, 2, 3 | 1, 2, 3 | |
| AF103 | Bacillus megaterium X | T, S | 96% (KC414697) | Firmicutes | 1, 2, 3 | 1, 2, 3 | 1, 2, 3 |
| AF102 | Bacillus subtilis X | T, S | 97% (KU551891) | Firmicutes | 1, 2, 3 | 1, 2, 3 | 1, 2, 3 |
| AF111 | Burkholderia cenocepacia | T, S | 96% (GU433447) | β Proteobacteria | 1, 2, 3 | ||
| AF129 | Burkholderia gladioli Z | S | 97% (EU1611873) | β Proteobacteria | 1, 2 | 2 | |
| AF109 | Chryseobacterium indologenes | T, S | 98% (KU551895) | Flavobacteria | 1, 2, 3 | 2 | |
| TL032 | Enterobacter aerogenes | T, S | 97% (AM184247) | γ Proteobacteria | 1 | 1, 2, 3 | |
| BT011 | Enterobacter spp. | T, S | 97% (KX518848) | γ Proteobacteria | 1 | ||
| TL012 | Flavobacterium johnsoniae | T, S | 96% (KU551897) | Flavobacteria | 1 | ||
| AF116 | Klebsiella oxytoca | T, S | 97% (KU551901) | γ Proteobacteria | 2 | ||
| AF128 | Klebsiella pneumoniae | T, S | 96% (KX518847) | γ Proteobacteria | 1 | ||
| TL015 | Pseduomonas hibiscola Y | T, S | 97% (KX518846) | γ Proteobacteria | 1, 2, 3 | 1, 2, 3 | |
| AF112 | Pseudomonas chlororaphis | T, S | 96% (KX518844) | γ Proteobacteria | 1, 2, 3 | ||
| AF115 | Pseudomonas geniculata X | T, S | 96% (KU551900) | γ Proteobacteria | 1, 2, 3 | 1, 2, 3 | 1, 2, 3 |
| TL011 | Pseudomonas lini | T, S | 99% (KX518842) | γ Proteobacteria | 1 | ||
| AF107 | Salmonella bongori | T, S | 96% (KU551899) | γ Proteobacteria | 1 | ||
| AF106 | Serratia marcescens | T, S | 96% (KX518843) | γ Proteobacteria | 1, 2, 3 | 1 | |
| TL010 | Sinorhizobium meliloti Y | T, S | 98% (KU551892) | α Proteobacteria | 1, 2, 3 | 1, 2, 3 | |
| TL009 | Stenotrophomas maltophilia Z | T, S | 98% (KU551894) | γ Proteobacteria | 1, 2, 3 | 1 | |
T= drought-tolerant maize line, S= drought-susceptible maize line.
TL= Tlaltizapán, Mor., AF= Agua Fría, Pue., EB= El Batán, Mex.
1=Autumn 2012; 2=Winter 2012; 3=Summer 2013.
X Highly frequent endophytes.
Y Frequent endophytes.
Z Low-frequent endophytes.
The taxonomic and functional structure of bacterial communities in soil is influenced by biotic and abiotic factors such as soil physical and chemical characteristics, weather conditions, plant genotype and interaction with other soil prokaryotes and eukaryotes, a fact that suggests that those interactions are complex. In this study, the structure and large number of isolated bacteria may be associated with the interaction of several factors, including plant genotype, the genetic characteristics of bacterium, soil, temperature, crop cycle and maize plant phenology (Li et al., 2014; Oliveira et al., 2009) that influence the colonization and dynamics of the endophytic bacterial community (Bodenhausen et al., 2013). A specific plant-endophyte relationship was revealed by performing bacterial chemotaxis on host plant exudates as carbon sources that act as signaling molecules (Albareda et al., 2006); it also showed that differences in exudate composition and patterns depend on the crop variety, development stage, exposure of the plant to stress and type of soil, which influence colonization by bacterial communities (Haichar et al., 2008). Studies on maize indicated that root exudates are 65% sugar, 33% organic acids and 2% amino acids, and that changes in the quantity and quality of exudation patterns at different root growth and physiological stages influenced the biomass and the structure of bacterial communities by increasing nutrient activity and deposition by those microbial communities and favored plant growth (Baudoin et al., 2003). Some maize studies that used a dependent culture approach have shown dynamic changes in the bacterial community of the rhizosphere at different crop growth stages (Cavaglieri et al., 2009; Nacamulli et al., 1997).
In this study, specific bacteria were identified such as Burkholderia gladioli, which was isolated at a low frequency only in susceptible maize lines at two locations and crop cycles (Table 1).
Burkholderia gladioli was previously identified in roots of a wild maize ancestor (Zea mays ssp. parviglumis) and in modern maize genotypes as an endophyte showing antifungal properties (Shehata et al., 2016). Also, Burkholderia sp. inoculation in maize had positive effects on drought tolerance (Fan et al., 2015; Naveed et al., 2014).
Density of endophytic populations in maize lines. The density of endophytic bacteria in the 14 maize lines ranged from 1, 6 Log10 UFC g-1 of root. The ANOVA showed highly significant differences (**= p≤0,01) in the density of the endophytic population of B. subtilis, B. megaterium, P. hibiscola and S. meliloti in terms of location, crop cycle, days after transplanting (dds) and group (drought tolerance/ susceptibility) of maize lines (Table 2).
Table 2 Variance analysis and comparison of means of population density (Log10 UFC g-1 of root) in roots of 14 drought-tolerant maize lines and drought-susceptible maize lines.
| B. subtilis | B. megaterium | P. hibiscola | S. meliloti | |
| Localidad | ** | ** | ** | ** |
| Ciclo | NS | ** | ** | NS |
| Días | NS | ** | NS | ** |
| Tolerancia | ** | ** | ** | ** |
| Tolerante | 2.2684 a | 3.7374 a | 2.9388 a | 4.8880 a |
| Susceptible | 1.7410 b | 3.3976 b | 2.4501 b | 4.6389 b |
Means with the same letter are not statistically different (DMS, 0.05).
NS= There are no significant differences.
**= Highly significant differences.
By location, the highest population densities recorded at El Batán were those of B. subtilis and P. hibiscola, while in Tlaltizapán they were B. megaterium and S. melilloti. By crop cycle and sampling date, B. megaterium and P. hibiscola were significantly different (p≤0,05) during the V-2012 cycle at 52 dds. In this study, a total of 22 endophytic bacteria were identified in roots of drought tolerant and drought susceptible maize lines. However, B. subtilis, B. megaterium, P hibiscola and S. meliloti population densities were significantly higher in drought tolerant maize lines than in susceptible lines (Table 2).
The density of bacterial populations is a key element, considered to be the greatest metabolism regulation mechanism in the interaction with the biotic and abiotic environment, through which it coordinates the expression of specialized genes, depending on cellular density. It has been demonstrated that this behavior, known as quorum sensing (QS) in bacteria associated with plants, regulates the expression of genes in the rhizosphere to synthesize secondary metabolites, antifungal compounds, antibiotics and extracellular enzymes involved in biocontrol (Somers et al., 2004; Whitehead et al., 2001).
In this study, the total population density of B. subtilis in the maize lines sown at El Batán was 3, 4 Log10 UFC g-1 of root, and there was no significant difference by cycle or days after sowing, but it was significantly higher in drought-tolerant maize lines (Table 2, Figure 1).

Figure 1 B. subtilis population in 7 drought-tolerant maize lines and 7 drought-susceptible maize lines at 25, 52 y 75 dds during the 2012 summer crop cycle, 2012 winter crop cycle and 2013 summer crop cycle in three locations of Mexico.
As an endophyte, B. subtilis is a microorganism of great interest in biotechnological applications as biocontrol agent for biotic and abiotic stresses because of its efficient colonization of plant roots which triggers different biocontrol and adaptation mechanisms in different environments (Marulanda et al., 2006). Additionally, this demonstrates its high capacity to produce volatile organic compounds (VOCs) that act as signaling molecules to trigger a defense response through specific induced systemic resistance (ISR) (Farag et al., 2013). These VOCs activate hormone production paths, including auxins, gibberellins, cytokinins and salicylic acid that foster host plant growth under stress conditions, mainly by increasing root biomass, which in turn improves water absorption (Zhang et al., 2007). Another study showed that maize seed inoculated with selected B. subtilis strains provided important benefits as a growth promoter and increased nutrient absorption capacity (Canbolat et al., 2006)In Tlaltizapán, the population density of B. megaterium in maize lines was from 3, 5Log10 UFC g-1 of root during the three cycles and significantly higher (5 Log10 UFC g-1 of root) in the V-2012 cycle at 52 dds, and in drought tolerant maize lines (Table 2, Figure 2).
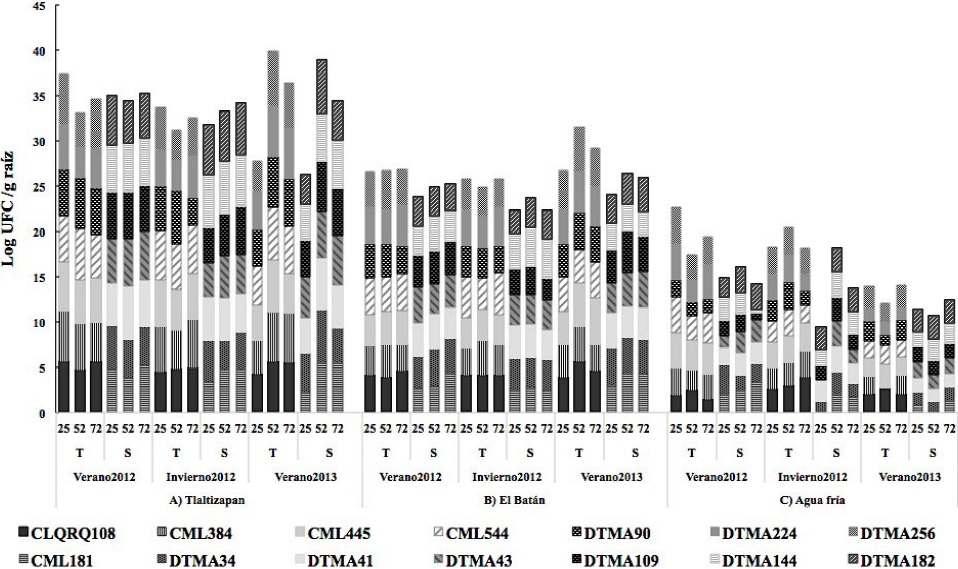
Figure 2 B. megaterium population in 7 drought-tolerant maize lines and 7 drought-susceptible maize lines at 25, 52 y 75 dds during the 2012 summer crop cycle, 2012 winter crop cycle and 2013 summer crop cycle in three locations of Mexico.
Recent studies have shown that B. megaterium modifies the maize plant response to several abiotic stresses, and the importance of the population density of some Bacillus species in the drought tolerance and water transportation response in Retama sphaerocarpa, because it promotes root growth and water absorption capacity, as well as the levels of proline and indoleacetic acid (Marulanda et al., 2006). In particular, those studies point out that the presence of B. megaterium in the host plant increases biomass and water content in the root (Marulanda et al., 2009), a fact that suggests that these endophytic populations could be used in maize plants sown in arid and semi-arid areas. The biological function of other endophytic Bacillus species in maize are related to the plant’s efficient defense response against pathogens associated with the production of antifungal lipopeptides that induces the expression of defense genes (Gond et al., 2015).
At El Batán, the population density of P. hibiscola in maize lines was from 3, 5 Log10 UFC g-1 of root during the three crop cycles and significantly higher (5 Log10 UFC g-1 of root) in the V-2012 cycle. It was not isolated in Agua Fría. There were no significant differences among the days after sowing (dds), but the density was significantly higher in drought-tolerant maize lines (Table 2, Figure 3).
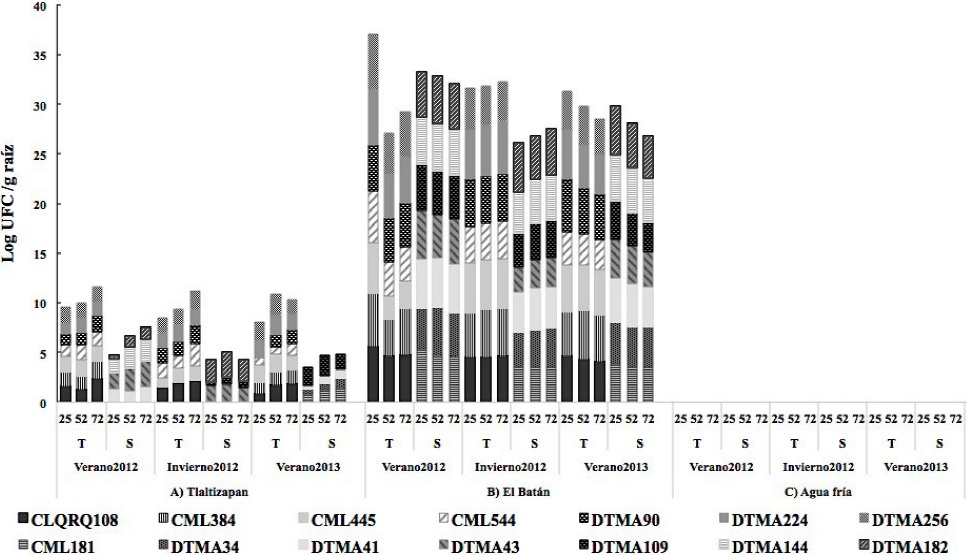
Figure 3 P. hibiscola population in 7 drought-tolerant maize lines and 7 drought-susceptible maize lines at 25, 52 y 75 dds during the 2012 summer crop cycle, 2012 winter crop cycle and 2013 summer crop cycle in three locations of Mexico.
P. hibiscola has not been cited as a maize endophyte. However, according to the phylogenetic affiliation of the P. hibiscola type ATCC 19867 strain based on a comparative analysis of 16S rADN gene sequences and chemical taxonomy profiles, it was reclassified as Stenotrophomonas sp. (Anzai et al., 2000). S. maltophilia has been cited as a maize endophyte (McInroy and Kloepper, 1995) and reported to act as a biological control agent against soil-borne pathogens such as Pythium spp., Fusarium spp. and Rhizoctonia solani. It has also been reported to produce indoleacetic acid, which regulates the development of sprouts and side roots in plants (Mehnaz et al., 2010).
The population density of S. meliloti in drought-tolerant maize lines in Tlaltizapán was from 4, 6 Log10 UFC g-1 of root during the three cycles, and significantly higher (6 Log10 UFC g-1 of root) at 52 dds. It was not isolated in Agua Fría. There were no significant differences per crop cycle, but it was significantly higher in drought-tolerant maize lines (Table 2, Figure 4).

Figure 4 S. meliloti population in 7 drought-tolerant maize lines and 7 drought-susceptible at 25, 52 y 75 dds during the 2012 summer crop cycle, 2012 winter crop cycle and 2013 summer crop cycle in three locations of Mexico.
S. meliloti is known to be a nitrogen-fixing nodular bacterium in plants of the Medicago genus, and involved in the drought tolerance and salinity response (Roumiansteva and Muntyan, 2015). In Mexico, the presence of a great genetic diversity of native S. meliloti populations in alfalfa (Medicago spp.) was reported, and so far, new species have been identified in woody legumes of the Leucaena (Wang et al., 2002) and Acacia genera (Toledo et al., 2003). The Sinorhizobium genus was found in the maize rhizosphere (Rosenblueth and Martínez-Romero, 2006), and Rhizobium etli bv. phaseoli as an endophyte in maize roots in Mexico (Gutiérrez-Zamora and Martínez-Romero, 2001). However, there is no information about S. meliloti as an endophyte in maize roots, so the present study provides reference information about the natural endophytic colonization of S. meliloti in maize in Mexico. Alfalfa and maize intercropping is common in the studied locations, so this may explain the endophytic colonization of S. meliloti in maize roots. Studies conducted on rice (Oryza sativa) suggest that S. meliloti may produce a lumichrome signaling molecule in the rhizosphere of those plants and promote growth by inducing better root respiration, stomatal conductance, leaf transpiration and photosynthetic efficiency (Chi et al., 2010). When it was applied at nanomolecular concentrations, it promoted legume and cereal growth and increased root biomass in the legume Lotus japonicus and in tomato (Solanum lycopersicum) (Gouws et al., 2012).
The frequency of certain bacteria genera isolated from maize roots in this study may be related to the phylogeny of the maize lines. For example, among bacteria showing higher frequency and density in drought-tolerant maize lines, two Bacillus species stand out. Bacillus is considered an important endophytic bacterium that has been isolated from both Teozintle (a maize ancestor) and modern maize genotypes; it has been shown that the composition of the endophytic bacteria colony in maize seed varies and that it has been preserved through the evolution, ethnography and ecology of the maize plant as a host (Johnston-Monje and Raizada, 2011).
In this research, we determined that some factors such as drought tolerance/susceptibility of the maize lines used, as well as the location, crop cycle and time (days after transplanting) significantly affected the density of cultivable endophytic bacteria in the roots of the maize genetic materials. Rhizobacteria as promoters of growth (PGPR), nutrition and plant disease management have been extensively studied, but their function in managing abiotic stresses such as drought has generated great interest in recent years (Dimpka et al., 2009; Grover et al., 2010; Kavamura et al., 2013; Yang et al., 2009). The use of endophytic microorganisms may be a viable alternative against biotic and abiotic stresses. Strains isolated in this study are a microbial resource that is intimately associated with maize and has the potential to be used as a biotechnological tool in agriculture; these strains deserve to be evaluated in the future as microbial inoculants and inductors of drought stress tolerance, as well as to other biotic and abiotic factors in regions of Mexico where maize is produced under limiting conditions.
Conclusions
Cultivable root endophytic bacteria that were most frequently present in the 14 maize lines tested belong to the Proteobacteria and Firmicutes phyla. The highest population density of endophytic bacteria was found in drought-tolerant maize lines. Bacteria of the Bacillus, Pseudomonas and Sinorhizobium genera showed the highest frequency and population density in drought-tolerant maize lines at the three study locations and crop cycles. Sinorhizobium meliloti is able to endophytically colonize maize roots.
Acknowledgments
The authors wish to thank the Colegio de Postgraduados and the Consejo Nacional de Ciencia y Tecnología (CONACyT) for funding this research and for the scholarship granted, as well as the International Maize and Wheat Improvement Center (CIMMYT) for providing the seed for conducting this research.
REFERENCES
Albareda M, Dardanelli MS, Sousa C, Megías M, Temprano F and Rodríguez D. 2006 Factors affecting the attachment of rhizospheric bacteria to bean and soybean roots. FEMS Microbiology Letters 259:67-73. https://doi.org/10.1111/j.1574-6968.2006.00244.x [ Links ]
Anzai Y, Kim H, Park JY, Wakabayashi H and Oyaizu H. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. International Journal of Systematic and Evolutionary Microbiology 50:1563-1589. DOI: 10.1099/00207713-50-4-1563 [ Links ]
Baudoin E, Benizri E and Guckert A., 2003. Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biology and Biochemistry 35:1183-1192. https://doi.org/10.1016/S0038-0717(03)00179-2 [ Links ]
Bodenhausen N, Horton MW and Bergelson J. 2013. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLOS ONE 8:e56329. https://doi.org/10.1371/journal.pone.0056329 [ Links ]
Canbolat M, Bilen S, Çakmakçi R, Sahin F and Aydi A. 2006. Effect of plant growth promoting bacteria and soil compaction on barley seeding growth, nutrient uptake, soil properties and rhizosphere microflora. Biology and Fertility of Soils 42:350-357. https://link.springer.com/article/10.1007/s00374-005-0034-9 [ Links ]
Cavaglieri L, Orlando J and Etcheverry M. 2009. Rhizosphere microbial community structure at different maize plant growth stages and root locations. Microbiological Research 164:391-399. https://doi.org/10.1016/j.micres.2007.03.006 [ Links ]
Chi F, Yang P, Han F, Jing Y and Shen S. 2010. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics 10:1861-1874. http://onlinelibrary.wiley.com/doi/10.1002/pmic.200900694/full [ Links ]
Cochran, W. G. Técnicas de muestreo. México: Compañía Editorial Continental, 1982. 513 p. [ Links ]
Dimpka C. Weinand T and Asch F. 2009. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell and Environment 32:1682-1694. DOI: 10.1111/j.1365-3040.2009.02028.x [ Links ]
Fan X, Hu H, Huang G, Huang F, Li Y and Palta J. 2015. Soil inoculation with Burkholderia sp. LD-11 has positive effect on water-use efficiency in inbred lines of maize. Plant Soil 390:337-349. https://doi.org/10.1007/s11104-015-2410-z [ Links ]
Farag MA, Zhang H and Ryu CM. 2013. Dynamic chemical communication between plants and bacteria through air-borne signals: induced resistance by bacterial volatiles. Journal of Chemical Ecology 39:1007-1018. https://link.springer.com/article/10.1007/s10886-013-0317-9 [ Links ]
FAO. 2016. Disponible en línea: http://www.fao.org/do-crep/003/x7650s/x7650s10.html (Consulta, marzo 2016) [ Links ]
FIRA, Fideicomisos Instituidos en Relación con la Agricultura. 2016. Disponible en línea: https://www.gob.mx/cms/uploads/attachment/file/61952/Panorama_Agroalimentario_Ma_z_2015.pdf (Consulta, febrero 2016) [ Links ]
Galkiewicz JP and Kellogg CA. 2008. Cross-Kingdom amplification using Bacteria-specific primers: complications for studies of coral microbial ecology. Applied Environmental Microbiology 74:7828-7831. Disponible en línea: http://aem.asm.org/content/74/24/7828.short [ Links ]
Gond SK, Bergen MS, Torres MS, White JF and Kharwar RF. 2015. Effect of bacterial endophyte on expression of defense in Indian popcorn against Fusarium moniliforme. Symbiosis 66:133-140. https://doi.org/10.1007/s13199-015-0348-9 [ Links ]
Gouws LM, Botes E and Wiese AJ, Trenkamp S, Torres-Jerez I, Tang J, Hills NP, Usadel B, Lloyd RJ, Fernie RA, Kossmann J and van der Merwe M. 2012. The plant growth promoting substance, lumichrome, mimics starch, and ethylene-associated symbiotic responses in lotus and tomato roots. Front Plant Science 120:1-20. https://dx.doi.org/10.3389%2Ffpls.2012.00120 [ Links ]
Grover M, Ali SZ, Sandhya V, Rasul A and Venkateswarlu B. 2010. Role of microorganisms in adaptation of agriculture crops to abiotic stresses. World Journal of Microbiology and Biotechnology 27:1231-1240. https://doi.org/10.1007/s11274-010-0572-7 [ Links ]
Gutiérrez-Zamora ML and Martinez-Romero E. 2001. Natural endophytic association between Rhizobium etli and maize (Zea mays L.). Journal of Biotechnology 91:117-126. https://doi.org/10.1016/S0168-1656(01)00332-7 [ Links ]
Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T and Achouak W. 2008. Plant host habitat and root exudates shape soil bacterial community structure. The ISME Journal 2:1221-1230. Disponible en línea: http://www.abdn.ac.uk/staffpages/uploads/mbi010/ISME%20Journal%2012,%201221-1230_1.pdf [ Links ]
Hardoim PR, Van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M and Sessitsch A. 2015. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiology and Molecular Biology Reviews 79:293-320. Disponible en línea: http://mmbr.asm.org/content/79/3/293.short [ Links ]
Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD and Lutzoni F. 2007. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Molecular Phylogenetics and Evolution 42:543-555. https://doi.org/10.1016/j.ym-pev.2006.07.012 [ Links ]
Ikeda CA, Bassani LL, Adamoski D, Stringari D, Cordeiro VK, Glienke C, Steffens MBR, Hungria M and Galli-Terasawa LV. 2013. Morphological and genetic characterization of endophytic bacteria isolated from roots of different maize genotypes. Microbial Ecology 65:154-160. DOI: 10.1007/ s00248-012-0104-0 [ Links ]
Johnston-Monje D and Raizada MN. 2011. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 6: e20396. https://doi.org/10.1371/journal.pone.0020396 [ Links ]
Kasim WA, Osman ME, Omar MN, Abd El-Daim IA, Bejai S and Meijer J. 2013. Control of drought stress in wheat using plant-growth- promoting bacteria. Journal of Plant Growth Regulation 32:122-130. Disponible en línea: https://link.springer.com/article/10.1007/s00344-012-9283-7 [ Links ]
Kamara YA, Menkir A, Badu-Apraku B and Ibikunle O. 2003. The influence of drought stress on growth, yield and yield components of selected maize genotypes. Journal of Agricultural Science 141:43-50. https://doi.org/10.1017/S0021859603003423 [ Links ]
Kavamura VN, Santos SN, Silva JL, Parma MM, Avila LA, Visconti A, Zucchi TD, Taketani RG, Andreote FD and Melo IS. 2013. Screening of Brazilian cacti rhizobacteria for plant growth promotion under drought. Microbiological Research 168:183-191. http://dx.doi.org/10.1016/j.micres.2012.12.002 [ Links ]
Kim YC, Glick BR., Bashan Y and Ryu CM. 2012. Enhancement of plant drought tolerance by microbes. In: Aroca R. (eds) Plant responses to drought stress. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-32653-0_15 [ Links ]
Li X, Rui J, Maoa Y, Yannarell A and Mackie R. 2014. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biology and Biochemistry 68: 392-401. https://doi.org/10.1016/j.soilbio.2013.10.017 [ Links ]
Liu Y, Zuo S, Zou YY, Wang JH and Song W. 2013. Investigation on diversity and population succession dynamics of endophytic bacteria from seeds of maize (Zea mays L., Nongda108) at different growth stages. Annals of Microbiology 63:71-79. Disponible en línea: https://link.springer.com/article/10.1007/s13213-012-0446-3 [ Links ]
Mahuku GS. 2004. A simple extraction method suitable for PCR based analysis of plant, fungal and bacterial DNA. Plant Molecular Biology Reporter 22:71-81. Disponible en línea: https://link.springer.com/article/10.1007%2FBF02773351?LI=true [ Links ]
Marulanda A, Barea JM and Azcón R. 2006. An indigenous drought- tolerant strain of Glomus intraradices associated with a native bacterium improves water transport and root development in Retama sphaerocarpa. Microbial Ecology 52:670-678. Disponible en línea: https://link.springer.com/article/10.1007/s00248-006-9078-0 [ Links ]
Marulanda A, Barea JM and Azcon R. 2009. Stimulation of plant growth and drought tolerance by native microorganisms (AM fungi and bacteria) from dry environments: mechanisms related to bacterial effectiveness. Journal of Plant Growth Regulation 28:115-124. Disponible en línea: https://link.springer.com/article/10.1007/s00344-009-9079-6 [ Links ]
McInroy JA and Kloepper JW. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant and Soil 173:337-342. Disponible en línea: https://link.springer.com/article/10.1007%2FBF00011472?LI=true [ Links ]
Mehnaz S, Kowalik T, Reynolds B and Lazarovits G. 2010. Growth promoting effects of corn (Zea mays) bacterial isolates under greenhouse and field conditions. Soil Biology and Biochemistry 42:1848-1856. https://doi.org/10.1016/j.soilbio.2010.07.003 [ Links ]
Montañez A, Blanco RA, Barlocco C, Beracochea M and Sicardi M. 2012. Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (Zea mays L.) and their inoculation effects in vitro. Applied Soil Ecology 58:21-28. https://doi.org/10.1016/j.apsoil.2012.02.009 [ Links ]
Morales Y, Juárez D, Aragón C, Mascarua M, Bustillos M, Fuentes L, Martínez R and Muñoz J. 2011. Growth response of maize plantlets inoculated with Enterobacter spp., as a model for alternative agriculture. Revista Argentina de Microbiología 43:287-293. Disponible en línea: http://www.scielo.org.ar/pdf/ram/v43n4/v43n4a09.pdf [ Links ]
Nacamulli C, Bevivino A, Dalmastri C, Tabacchioni S and Chiarini L. 1997. Perturbation of maize rhizosphere microflora following seed bacterization with Burkholderia cepacia MCI 7. FEMS Microbiology Ecology. 23:183-193: https://doi.org/10.1111/j.1574-6941.1997.tb00401.x [ Links ]
Naseem H and Bano A. 2014. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance in maize. Journal of Plant Interactions. 9:689-701. https://doi.org/10.1080/17429145.2014.902125 [ Links ]
Naveed M. Mitter B, Reichenauer TG, Wieczorek K and Sessitsch A. 2014. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp. FD 17. Environmental and Experimental Botany 97:30-39. https://doi.org/10.1016/j.envexpbot.2013.09.014 [ Links ]
Oliveira ALM, Stoels M, Schmid M, Reis VM, Baldani JI and Hartmann A. 2009. Colonization of sugarcane plantlets by mixed inoculations with diazotrophic bacteria. European Journal of Soil Biology 45:106-113. https://doi.org/10.1016/j.ejsobi.2008.09.004 [ Links ]
Pereira P, Ibáñez F, Rosenblueth M, Etcheverry M and Martínez-Romero E. 2011. Analysis of the bacterial diversity associated with the roots of maize (Zea mays L.) through culture-dependent and culture-independent methods. International Scholarly Research Network 2011:1-10. DOI: 10.5402/2011/938546 [ Links ]
Rai R, Prasanta K, Dash BM, Prasanna AS. 2007. Endophytic bacterial flora in the stem tissue of a tropical maize (Zea mays L.) genotype: isolation, identification and enumeration. World Journal Microbiology and Biotechnology 23:853-858. DOI:10.1007/s11274-006-9309-z [ Links ]
Rosenblueth M and Martinez-Romero E. 2006. Bacterial endophytes and their interaction with hosts. Molecular Plant-Microbe Interactions 19:827-837. Disponible en línea: http://apsjournals.apsnet.org/doi/pdf/10.1094/MPMI-19-0827 [ Links ]
Roumiansteva ML and Muntyan VS. 2015. Root nodule bacteria Sinorhizobium meliloti: Tolerance to salinity and bacterial genetic determinants. Microbiology 84:303-318. DOI: 10.1134/S0026261715030170 [ Links ]
Saravanakumar D, Kavino M, Raguchander T, Subbian P and Samiyappan R. 2011. Plant growth promoting bacteria enhance water stress resistance in green gram plants. Acta Physiology Plant 33:203-209. Disponible en línea: https://link.springer.com/content/pdf/10.1007%2Fs11738-010-0539-1.pdf [ Links ]
Shehata HR, Lyons EM, Jordan KS and Raizada MN. 2016. Bacterial endophytes from wild and ancient maize are able to suppress the fungal pathogen Sclerotinia homoeocarpa. Journal of Applied Microbiology 120:756-769. Disponible en línea: http://onlinelibrary.wiley.com/doi/10.1111/jam.13050/epdf [ Links ]
Somers E, Vanderleyden J and Srinivasan M. 2004. Rhizosphere bacterial signalling: A love parade beneath our feet. Critical Reviews in Microbiology: 30:205-240. DOI: 10.1080/10408410490468786 [ Links ]
Timmusk S, Islam A, Abd El D, Lucian C, Tanilas T and Kannaste A, Behers L, Nevo E, Seisenbaeva G, Stenströ E and Niinemets Ü. 2014. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS One 9:1-13. https://doi.org/10.1371/journal.pone.0096086 [ Links ]
Toledo I, Lloret L and Martínez-Romero E. 2003. Sinorhizobium americanum sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Systematic and Applied Microbiology 26:54-64. Disponible en línea: http://www.sciencedirect.com/science/article/pii/S0723202004701599 [ Links ]
Wang E, Tan ZY, Willems A, Fernández-López M, Reinhold-Hurek B and Martínez-Romero E. 2002. Sinorhizobium morelense sp. nov., a Leucaena leucocephala-associated bacterium that is highly resistant to multiple antibiotics. International Journal of Systematic and Evolutionary Microbiology 52:1687-1693. http://dx.doi.org/10.1099/00207713-52-5-1687 [ Links ]
Wilkinson S and Davies WJ. 2010. Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant, Cell & Environment 33:510-525. Disponible en línea: http://onlinelibrary.wiley.com/doi/10.1111/j.1365-3040.2009.02052.x/full [ Links ]
Whitehead AN, Barnard LMA, Slater H, Natalie JL, Simpson G and Salmond PC .2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiology Reviews 25:365-404. https://doi.org/10.1111/j.1574-6976.2001.tb00583.x [ Links ]
Yang J, Kloepper JW and Ryu C. 2009. Rhizosphere bacteria help plants tolerate abiotic stress. Trends in Plant Science 14:1-4. https://doi.org/10.1016/j.tplants.2008.10.004 [ Links ]
Yasmin H, Bano A and Samiullah A. 2013. Screening of PGPR isolates from semi-arid region and their implication to alleviate drought stress. Pakistan Journal Botany 45: 51-58. Disponible en línea: https://www.scopus.com/record/display.uri?eid=2-s2.0-84873424619&origin=inward&txGid=f67ad4b109d3d368731a8b22a1f14cfa [ Links ]
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS and Pare PW. 2007. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839-851. Disponible línea: https://link.springer.com/content/pdf/10.1007%2Fs00425-007-0530-2.pdf [ Links ]
Received: October 10, 2017; Accepted: December 07, 2017











 text in
text in 


