Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.36 n.1 Texcoco Jan./Apr. 2018
https://doi.org/10.18781/r.mex.fit.1709-2
Scientific articles
Detection of Sugarcane mosaic virus (SCMV) in Saccharum spp. in Mexico and phylogenetic origin of one isolate from Jalisco
1 Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias. Campo Experimental Tecomán, Laboratorio de Biotecnología de Plantas, Km 35 carretera Colima-Manzanillo, CP. 28100, Tecomán-Colima.
2 Instituto Tecnológico de Colima. Avenida Tecnológico No. 1, CP. 28976, Villa de Álvarez, Colima.
3 Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias. Campo Experimental Tecomán. Km 35 carretera Colima-Manzanilla, CP. 28100, Tecomán-Colima.
4 Instituto Tecnológico de Tlajomulco. Km 10 carretera Tlajomulco-San Miguel Cuyutlán, CP. 45640, Tlajomulco de Zúñiga, Jalisco.
Sugarcane mosaic virus (SCMV) is one of the main viral agents that infect sugarcane (Saccharum spp.). SCMV detection in Mexico has been based on typical symptomatology of the disease, which is not conclusive. Additionally, there is limited information about their phylogenetic origin. The objective of this work was to detect the presence and distribution of SCMV in the sugarcane growing areas of the Mexican Pacific using RT-PCR, and to determine the phylogenetic origin of one isolate from Jalisco. The results showed the wide distribution of SCMV in sugarcane areas of the Mexican Pacific. The virus was found in 33 of the 242 samples analyzed, corresponding to 13.63%. The varieties Mex 69-290 and CP 72-2086 presented the most severe mosaic symptoms. Phylogenetic analysis using partial HC-Pro sequences of SCMV isolate from Jalisco (JalMex-126) suggests a close relationship with isolates from India, Australia, China and Argentina, and thus likely share a common genetic origin and have been dispersed to through infected sugarcane germplasm.
Key words: sugarcane; virus; RNA; phylogeny
El virus de mosaico de la caña de azúcar (SCMV) es uno de los principales agentes virales que infectan a caña de azúcar (Saccharum spp.). En México la detección del SCMV se ha basado en sintomatología típica de la enfermedad, por lo que la información no es concluyente, además hay poca información sobre su origen filogenético. El objetivo del trabajo fue detectar mediante RT-PCR la presencia y distribución del SCMV en las zonas cañeras de los estados de Colima, Jalisco y Nayarit, México y determinar el origen filogenético de un aislado de Jalisco. Los resultados obtenidos demuestran la amplia distribución del SCMV en la región cañera del Pacífico de México. De las 242 muestras analizadas se detectó al virus en 33 de ellas, lo que corresponde a un 13.63%. Las variedades Mex 69-290 y CP 72-2086 fueron las que presentaron los síntomas de mosaico más severos. El análisis filogenético con la secuencia parcial HC-Pro del SCMV aislado de Jalisco (JalMex-126) sugiere una estrecha relación con aislados de la India, Australia, China y Argentina, por lo que probblemente compartan un origen genético común y se hayan dispersado a través de germoplasma de caña de azúcar infectado.
Palabras clave: caña de azúcar; virus; RNA; filogenia
Introduction
Diseases caused by phytopathogens are an important factor for sugarcane production. In Mexico, approximately 80% of the diseases that affect this crop are fungal in origin, and the remaining 20% is caused by bacteria, viruses and phytoplasmas (CONADESUCA, 2015). Some of the first epidemiologies on the sugarcane crop reported in the world in the beginning of the 20th Century include those caused by viruses, causing large economic losses (Grisham, 2000). The Sugarcane mosaic virus (SCMV), causal agent of the mosaic disease, is one of the most economically important causal agents of the mosaic disease worldwide (Gonçalves et al., 2012). The disease caused drastic outbreaks in Argentina, Brazil, Cuba, Puerto Rico, and the United States (Koike and Guillaspie, 1989; Yang and Mirkov, 1997). This lead to the introduction of interspecific hybrids of the genus Saccharum (mosaic-tolerant) imported from Java, in order to control the rapid spread of the disease in noble canes (S. officinarum) obtained back then in those countries (Koike and Gillaspie, 1989). In susceptible varieties infected with SCMV, yield losses are estimated in 11-50% (Singh et al., 2003; Singh et al., 2005). In Mexico, the first report (based on symptoms) of this virus affecting sugarcane was carried out in 1929, in El Potrero, Veracruz. In 1947, over 80% of the areas planted with native sugarcane varieties, from the Independencia Mill in the municipal area of Martínez de la Torre, presented the disease (CONADESUCA, 2015). In that same country, the first report (based on molecular methods) of SCMV affecting maize plants was carried out in 2006 (Espejel et al., 2006), although there is no quantitative information on the damages this disease causes in sugarcane plantations in Mexico.
The general symptom caused by SCMV is characterized mostly by the paling of the leaf blade, which presents normally green colored areas, alternated with pale or yellow-green areas; these areas are a result of the variations in the levels of chlorophyll in the leaf (Grisham, 2000; CONADESUCA, 2015). The symptoms of this disease may also be caused by other viruses and may be confused with the Sugarcane streak mosaic virus (SCSMV) or the Sorghum mosaic virus (SrMV), which also affect sugarcane plants (Viswanathan et al., 2008; Xie et al., 2009). Asymptomatic plants can also be positive for SCMV (Xu et al., 2008). In Mexico, reports on the detection of SCMV by molecular methods are scarce, and have mainly been based on symptoms (CONADESUCA 2015), so there is no certainty that the visual damage of the mosaic truly corresponds to SCMV or any other virus that presents similar symptoms. Several reports have standardized protocols to detect SCMV using RT-PCR (Smith and Van de Velde, 1994; Xie et al, 2009; Filippone et al., 2010). Smith and Van de Velde (1994) developed primers S400-551 and S400-910, which amplified a 359 bp fragment, which correspond to a partial region that codifies for the capsid protein (CP) of SCMV. In another study, Yang and Mirkov (1997) developed the primers SCMV-F3/SCMV-R3, which amplify a roughly 900 bp band, which corresponds to a region that codifies for the CP protein of SCMV. Xie et al. (2009) designed the set of oligonucleotides SCMV-F1/SCMV-R1 based on sequences deposited in the NCBI database; these primers amplified a 720 bp fragment, which corresponds to a partial region of the HC-Pro protein of SCMV. Recently, Filippone et al. (2010) optimized a protocol to detect SCMV using the oligonucleotides described earlier by Yang by Mirkov (1997) and Smith and Van de Velde (1994), respectively.
On the other hand, it is important to know the origin of SCMV in order to know possible entries of this disease into Mexico. In the year 2012, a phylogenetic analysis was reported, with 185 SCMV CP sequences from several countries, suggesting different phylogeographic origins of two Mexican isolates that this study considered (Chaves and Ortiz, 2012). On the other hand, the results produced by Xie et al. (2016) on the phylogenetic analysis of 24 SCMV isolates from China and other countries revealed that they could be divided into two groups, which were related to the SCMV host plant species. More recently, results by Moradi et al. (2017) from CP sequences of SCMV from various countries suggest five divergent evolutionary lineages, where the geographic origin and/or SCMV host plants are partially related. Due to this, the aim of this study is to detect, using RT-PCR, the presence and distribution of the SCMV in the sugarcane producing areas of the Mexican Pacific, and to establish the phylogenetic origin of one isolate from the state of Jalisco. The information produced in this work may be used by the Sugarcane Reasearch and Development Center (Centro de Investigación y Desarrollo de la Caña de Azúcar, CIDCA), both in its quarantine station for the exchange of germplasm, and in the genetic breeding program, in which the selection and elimination of clones susceptible to SCMV are routine procedures.
Materials and methods
Plant material
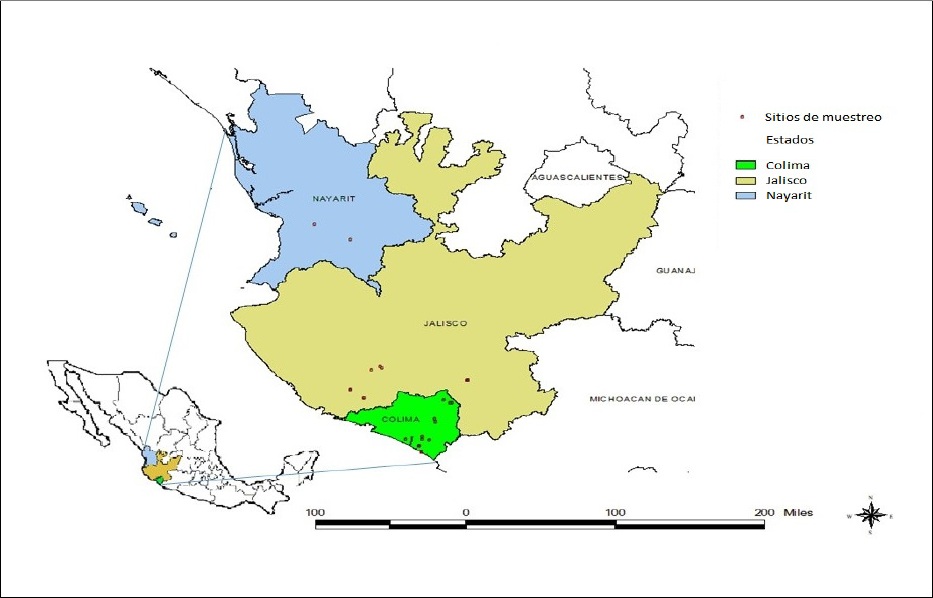
In the year 2014, foliar samples were collected from diverse varieties of sugarcane planted commercially in the states of Colima, Jalisco and Nayarit (Figure 1). Plants displayed typical mosaic symptoms or they were asymptomatic. The samples taken were wrapped in plastic bags, placed in coolers with thermal refrigerants and were transferred to the Laboratory of Plant Biotechnology from Campo Experimental Tecomán of INIFAP, located on km 35 of the Colima-Manzanillo highway, in Colima.
Extraction and quantification of the total RNA
Approximately 200 mg were taken from several parts of the foliar tissue of one same plant. These leaves were pulverized using liquid nitrogen until a fine powder was formed, which was deposited in a 1.5 mL tube, and homogenized with 500 μL of the Tripure reagent (Roche). Manufacturer recommendations were taken for the extraction of total RNA. Finally, the RNA obtained was resuspended in RNase free water (0.01% DEPC) and stored at -70°C. Quantification was carried out using a NanoDrop (Thermo Scientific) spectrophotometer using 1 μL of the total RNA extracted, and the A260:280 y A260:230, ratio was measured to determine its purity.
Cloning a fragment of SCMV in pGEM-T Easy
In order to have a positive control for the detection of the virus, the PCR product of 720 bp, which corresponds to a sugarcane foliage sample, obtained in a location of the state of Jalisco, was purified from agarose gel using the “QIAquick Gel Extraction” (QIAGEN) kit, following manufacturer instructions. Later, the fragment was cloned using the “pGEM-T Easy” (Promega) system, and was transformed into E. coli JM109 cells (Promega), which were previously made competent using CaCl2 according to Riley et al., 2008. A DNA fragment insert cloned in the vector was verified by purification of plasmid DNA from the recombinant cells and the subsequent digestion reaction with enzymes EcoR I and Not I.
Analysis of sequences
The plasmid from the DNA fragment cloned in E. coli was sequenced in a ABI PRISM 310 Genetic Analyzer in both strands with the method of termination with Applied Biosystems’ Big Dye. The editing and assembling of the “forward” and “reverse” sequences was carried out using the program CLC Main Workbench 7. Finally, the similarity of the sequences obtained was compared with those reported for SCMV in the National Center for Biotechnology Information (NCBI) database, using the Basic Local Alignment Search Tool (BLAST).
Detection of SCMV by RT-PCR
The reverse transcription reaction (RT) of the total RNA was carried out using the Reverse Transcription System (Promega) kit. The RNA was denaturalized at 70°C for 10 min; the RT reaction mixture was carried out in a volume of 20 µL containing 5 mM of MgCl2, buffer RT 1 X, 1 mM of each dNTP, 1 u/µL of recombinant RNasin® ribonuclease inhibitor, 15 u/µg of AMV reverse transcriptase AMV, 0.5 µg of oligonucleotide mixture by µg of RNA and 5 µL of total RNA (1.5 µg approximately). This mixture was incubated at room temperature for 10 min and then at 42°C for 45 min. Immediately, it was incubated at 95°C for 5 min, and finally 0-5°C for 5 min. The resulting cDNA were used as a mold for the amplification by PCR with the oligonucleotides described by Xie et al. (2009). The final volume of the reaction mixture was 25 µL, containing 12.5 µL of REDTaq® ReadyMixTM (SIGMA-ALDRICH), 1 μM of each oligonucleotide, 3 µL of cDNA and molecular grade water. The reaction mixture was incubated in a thermocycler (Labnet) with the following program: one cycle of 50°C for 30 min and 94°C for 2 min; 35 cycles of 94°C for 30 sec, 56°C for 30 sec and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Electrophoresis was carried out on agarose gel at 1% with TAE buffer 1 X and 12.5 µL of the PCR products were used; the samples were run with a voltage of 120 V. Finally, the gels were stained with BrEt and observed using a transilluminator with UV light (UVP) for the analysis of results.
Phylogenetic analysis
All bioinformatic analysis were carried out using CLC Workbench software version 7.0.3. Sequences of the BLAST analysis were selected and downloaded in NCBI with the highest percentages of identity with the SCMV sequence, wich named JalMex-126 (KT334297) and others from the same database were used (Table 2). For all samples, PCR in silico were carried out with the primers for SCMV described by Xie et al. (2009). In all the sequences in which 720 bp fragments were produced, these were extracted and stored in a local database. The sequences that did not generate the expected fragment were aligned with those generated in the database. The aligned regions of these sequences were selected and stored. The region HC-Pro of PRSV (NC_001785) was used as an external group. Multiple alignments of sequences were carried out using the MUSCLE algorithm, and the maximum likelihood and Neighbor-joining methods were used for the construction of the phylogenetic tree. General Time Reversible (GTR) was used as a model of substitution of nucleotides. The bootstrap analysis was carried out with 1,000 repetitions, and finally, the phylogenetic tree was edited using the software TreeGraph 2.
Table 1 List of sugarcane foliage samples that tested positive for SCMV by RT-PCR in the sugarcane-producing region of the Mexican Pacific.
| No. | Muestra | Estado | Municipio | Georeferencia | Variedad |
| 1 | 87 | Colima | Colima | N 19°11’33.0’’ W 103°43’15.4’’ | CP 72-2086 |
| 2 | 88 | Colima | Colima | N 19°09’43.6’’ W 103°43’14.0’’ | CP 72-2086 |
| 3 | 35 | Colima | Cuauhtémoc | N 19°24’34.14’’ W 103°38’19.79’’ | Mex 69-290 |
| 4 | 82 | Colima | Cuauhtémoc | N 19°22’36.61’’ W 103°33’37.49’’ | CP 72-2086 |
| 5 | 90 | Colima | Cuauhtémoc | N 19°22’09.12’’ W 103°34’49.81’’ | CP 72-2086 |
| 6 | 148 | Colima | Tecomán | N 18°52’36.24” W 103°52’1.49” | CP 72-2086 |
| 7 | 336 | Colima | Tecomán | N 18°52’40.12” W 103°51’50.62” | CP 72-2086 |
| 8 | 23 | Colima | Tecomán | N 18°56’37.4’’ W 103°46’26.03’’ | Mex 69-290 |
| 9 | 33 | Colima | Tecomán | N 18°48’33.23’’ W 103°50’37.89’’ | Mex 69-290 |
| 10 | 146 | Colima | Tecomán | N 18°57’57.26’’ W 103°50’22.01’’ | Saccharum spp. |
| 11 | 2 | Colima | Tecomán | N 18°56’06.52’’ W 104°00’03.54’’ | Saccharum spp. |
| 12 | 37 | Colima | Tecomán | N 18°48’32.91’’ W 103°50’33.48’’ | Mex 69-290 |
| 13 | 133 | Colima | Tecomán | N 18°52’37.61” W 103°51’57.66” | Saccharum spp. |
| 14 | 387 | Colima | Tecomán | N 18°57’56.03’’ W 103°50’22.02’’ | Saccharum spp. |
| 15 | 350 | Jalisco | Autlán | N 19°45’07.23’’ W 104°19’36.69’’ | Mex 69-290 |
| 16 | 342 | Jalisco | Cuautitlán | N 19°26’49.58’’ W 104°24’10.45’’ | Saccharum spp. |
| 17 | 344 | Jalisco | Cuautitlán | N 19°26’54.74’’ W 104°23’40.70’’ | Mex 79-431 |
| 18 | 355 | Jalisco | El Grullo | N 19°48’49.81’’ W 104°14’35.95’’ | Mex 69-290 |
| 19 | 349 | Jalisco | El Grullo | N 19°47’44.20” W 104°13’38.63” | Atemex 96-40 |
| 20 | 413 | Jalisco | La Huerta | N 19°31’07.39’’ W 104°32’11.61’’ | Saccharum spp. |
| 21 | 341 | Jalisco | La Huerta | N 19°31’08.14’’ W 104°32’11.61’’ | Saccharum spp. |
| 22 | 369 | Jalisco | La Huerta | N 19°31’11.23’’ W 104°32’10.97’’ | Saccharum spp. |
| 23 | 340 | Jalisco | La Huerta | N 19°31’05.15’’ W 104°32’06.51’’ | Atemex 96-40 |
| 24 | 114 | Jalisco | Zapotiltic | N 19°38’43.00’’ W 103°24’31.01’’ | Atemex 96-40 |
| 25 | 120 | Jalisco | Zapotiltic | N 19°38’42.90’’ W 103°24’29.40’’ | Atemex 96-40 |
| 26 | 125 | Jalisco | Zapotiltic | N 19°38’24.12’’ W 103°25’03.64’’ | CP 72-2086 |
| 27 | 110 | Jalisco | Zapotiltic | N 19°38’27.25’’ W 103°25’04.85’’ | CP 72-2086 |
| 28 | 113 | Jalisco | Zapotiltic | N 19°38’42.9’’ W 103°24’29.7’’ | Atemex 96-40 |
| 29 | 126 | Jalisco | Zapotiltic | N 19°38’42.68’’ W 103°24’30.04’’ | Atemex 96-40 |
| 30 | 123 | Jalisco | Zapotiltic | N 19°38’42.31’’ W 103°24’31.21’’ | Atemex 96-40 |
| 31 | 138 | Jalisco | Zapotiltic | N 19°38’42.58’’ W 103°24’31’’ | Atemex 96-40 |
| 32 | 186 | Nayarit | Santa María del Oro | N 21°17’56.3’’ W 104°32’14.9’’ | Mex 69-290 |
| 33 | 170 | Nayarit | Xalisco | N 21°27’50.1’’ W 104°53’11.4’’ | Saccharum spp. |
Table 2 Partial sequences of the fragment HC-Pro of SCMV obtained from the NCBI database and used in this study for the phylogenetic analysis.
| No. | Nombre del aislado | Origen | Hospedero | Año | No. Acceso |
| 1 | JalMex-126 | México | Saccharum spp. (Var. Atemex 96-40) | 2015 | KT334297 |
| 2 | Brisbane | Australia | Saccharum spp. | 2005 | AJ278405 |
| 3 | FZ-C1 | China | Saccharum spp. (Var. Badila) | 2014 | KR108212 |
| 4 | CBTCT-Seng | India | Saccharum sinense | 2014 | KX266877 |
| 5 | ARG-915 | Argentina | Saccharum spp. | 2007 | JX237863 |
| 6 | FZ-C2 | China | Saccharum spp. (Var. Badila) | 2014 | KR108213 |
| 7 | CBMungo | India | Saccharum barberi | 2014 | KX266876 |
| 8 | JAL-1 | México | Zea mays | 2010 | GU474635 |
| 9 | SCMV-VER1 | México | Zea mays | 2011 | EU091075 |
| 10 | Seehausen | Alemania | - | 2012 | JX185303 |
| 11 | SCMVgp1 | China | Zea mays | 2015 | NC_003398 |
Results
Symptoms
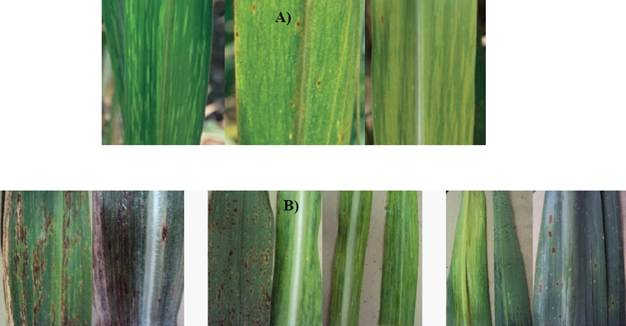
The symptoms related to the presence of SCMV were found in the foliar samples collected from sugarcane plants of different varieties in the states of Colima, Jalisco, and Nayarit, Mexico. Also, variations were observed in the mosaic patterns (Figure 2 A). Other samples with mosaics were found to have symptoms, such as white stripes in ribs, lesions from pustules, chlorosis, necrosis and corrugated leaves (Figure 2 B).
Cloning a fragment of SCMV
The positive control used to detect the presence and distribution of SCMV in three states of the Mexican Pacific was obtained from a sample with severe mosaic symptoms from the state of Jalisco. A 720 bp fragment was amplified and cloned; later, seven recombinant colonies were obtained and a restrictive analysis was carried out with EcoR I and Not I to verify the presence of the cloned fragment (Figure 3), which was finally sequenced. The BLAST analysis of the sequence presented an identity of up to 95% with other SCMV accessions, and was placed in the NCBI database, under registration number KT334297. The sequence was established as belonging to a partial region of the HC-Pro gene of SCMV.
Identification and distribution of SCMV in three states of the Mexican Pacific
All the products of RT-PCR that underwent electrophoresis in agarose gel that showed the amplification of fragments of 720 bp of SCMV with the set of oligonucleotides SCMV-F1/ SCMV-R1 described by Xie et al. (2009) were considered as positive to the virus (Figure 4). A total of 242 foliar samples of diverse, commercially planted, symptomatic and asymptomatic sugarcane varieties, were analyzed for mosaic, as well as other symptoms described earlier.
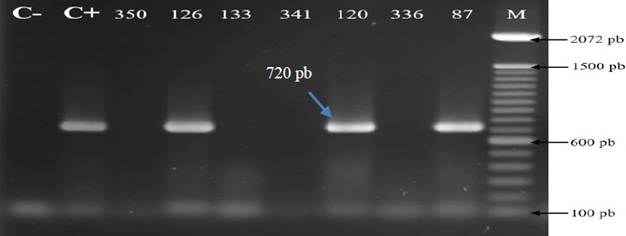
Figure 4 Detection of the Sugarcane mosaic virus (SCMV) by RT-PCR in sugarcane samples from three states of the Mexican Pacific. C-: negative control (H2O). C+: Positive control (plasmid DNA). M: Molecular weight marker 100 pb (Invitrogen). Numbers correspond to different samples analyzed.
Table 1 shows the wide distribution presented by SCMV in the tropical and subtropical regions of the Mexican Pacific covered by the present study. In the state of Colima, it was found in the varieties Mex 69-290 and CP 72-2086, among many others in the experimental stage. Also, in Jalisco it affected the same two varieties, as well as Atemex 96-40. Finally, in Nayarit variety Mex 69-290 was also the most affected. Table 1 shows the 33 sugarcane plants that tested positive for SCMV and their geographic origin, corresponding to 13.63% of the total of plants analyzed. The varieties with the most severe viral symptoms and that tested positive for SCMV were Mex 69-290, CP 72-2086 and Atemex 96-40. Some samples presented clear mosaic symptoms, although they tested negative for the presence of the virus by RT-PCR.
Phylogenetic origin
The phylogenetic analysis with the partial sequence HC-Pro of SCMV from Jalisco and other similar sequences from other countries is shown in Figure 5. The formation of two groups is observed, the first of which includes two sequences from Mexico, EU091075 and GU474635, whose percentages of identity with JalMex-126 (isolate from this study) were 80.39 and 79.28%, respectively. This group also included another two sequences from China and Germany. In the second group is JalMex-126, along with two sequences from India, one from Australia, two from China, and one from Argentina, with which it shares identity percentages of between 92.5-95.14%, and therefore the viruses of this group may have a common genetic origin. For both groups, the geographic origins of the isolates were very diverse. The main difference between these two groups formed lies in the type of virus host. The viral sequences of the first group had maize plants as hosts, except for the isolate from Germany, whose host is unknown. The rest of the HC-Pro sequences of SCMV that made up the second group come from diverse hybrids and sugarcane species (Table 2).

Figure 5 Phylogenetic tree based on the Neighbor-joining and maximum likelihood methods using partial sequences of the fragment HC-Pro of SCMV. Branches with bootstrap values below 50% were collapsed. The Papaya ringspot virus (PRSV) was used as an external group. The isolate of this study is highlighted in bold letters and underlined.
Discussion
The variety of symptoms present in the analyzed samples, combined with mosaic patterns, are possibly due to mixed infections with fungi, since some of the symptoms observed are from diseases such as brown spot, common rust, pokkah boeng, the causal agents of which are Cercospora longipes, Puccinia melanocephala and Gibberella fujikuroi, respectively (Raid and Comstock, 2000; Saumtally and Sullivan, 2000; Whittle and Irawan, 2000). However, molecular tests must be carried out to confirm the identity of these causal agents. On the other hand, according to Xie et al. (2009), the mosaic disease is caused by a complex of three viruses: the SCMV, the Sorghum mosaic virus (SrMV) and the Sugarcane streak mosaic virus (SCSMV), the last two of which have not been reported as affecting sugarcane in Mexico. The fact that samples with mosaic characteristics turned out negative for the RT-PCR test for SCMV in this study may be due to its symptoms being related with the presence of some of the viruses mentioned earlier (Grisham, 1994; Xie et al., 2009), or those samples with severe mosaics and positive to SCMV may contain mixed viral infections. Recently, Balarabe et al. (2014) mentioned that the Johnsongrass mosaic virus (JGMV) and the Maize dwarf mosaic virus (MDMV) cause mosaic disease in sugarcane in Nigeria. To this day, the presence of the SrMV has been confirmed in the United States, China, and Vietnam (Yang and Mirkov, 1997; Grisham and Pan, 2007; Ha et al., 2008; Zhang et al., 2016), whereas SCSMV is limited to the sugarcane producing areas of the Asian continent (Hall et al., 1998; Hema et al., 1999; Rao et al., 2006). However, due to the proximity of Mexico to the United States, it is likely that SrMV is infecting sugarcane plants in our country.
The detection of SCMV by RT-PCR in this study confirms its wide distribution in the sugarcane growing areas of the Mexican Pacific region. The virus is even present in the European continent, due to the amount of plant species that can act as reservoirs for it (Dallwitz, 1980; Dallwitz et al., 1993; Brunt et al., 1996). In Mexico, varieties Mex 69-290 and CP-72-2086, not only cover 50% of the area planted with sugarcane, but have also been in use for over 30 years, which has led to the genetic deterioration of the materials, leading, with time, to susceptibility to several diseases, including viral diseases. Also, its vegetative reproduction characteristics (seed-stake), as well as the presence of vector aphids Melanaphis sacchari and Rophalosiphum maidis (Figueredo et al., 2004) have promoted the dispersion of the virus.
Several reports have identified the presence of SCMV in various countries (Smith and Van de Velde, 1994; Xie et al, 2009; Filippone et al., 2010; Sawazaki et al., 2013). This study was based on the work described by Xie et al. (2009) for the standardization of the method of detection for RT-PCR. However, despite the molecular diagnosis techniques being more sensitive than serological methods, the latter are less expensive and easier to use when a higher number of samples must be processed (Gonçalves et al., 2007; Balarabe et al., 2014). In Mexico, the SCMV is viewed as the only causal agent of the mosaic disease, although there are no solid reports or with scientific validity to sustain these facts, since information on the presence of the virus in the country go back to 1930-1950, and have also been based on the typical symptoms of the disease (CONADESUCA, 2015), which is not conclusive. Studies related to the SCMV in Mexico have focused on maize (Espejel et al., 2006; Chaves et al., 2011; Chaves and Ortiz, 2012). Chaves et al. (2011) obtaine done isolate of SCMV in maize in the state of Veracruz and they used the sugarcane varieties CP 72-2086 and MY 44-12 as reference genotypes to inoculate the virus in the leaf of both maize and sugarcane to determine the movement of SCMV-Ver1. Due to this, the present paper represents the first study focused on sugarcane to determine the presence and distribution of SCMV in the sugarcane producing area of the Mexican Pacific. Finally, it is worth pointing out that in Mexico, there is a need for more studies that quantify the damage caused by the sugarcane mosaic disease, since most farmers or the agroindustry are unaware of the economic impact it could have on the crop.
Regarding the phylogenetic origin of the isolation JalMex-126, results indicate that it could have a common genetic origin with isolations from India, Australia, China and Argentina, which suggests deficiencies in the phytosanitary control during the exchange of germplasm, which is very common due to the type of seed-stake reproduction in sugarcane. However, due to the number of HC-Pro sequences of SCMV, it is very reduced in the NCBI data base in comparison with the CP sequences, it was not possible to relate the origin of the Mexican isolation with important sugarcane producing countries such as Brazil, United States, Colombia, and others. In this study we obtained the formation of two groups based on partial HC-Pro sequences of SCMV, and this same tendency was previously reported by Chaves and Ortiz (2012) and Xie et al. (2016), using CP sequences of SCMV. Similarly, these authors coincide in that the formation of these groups is related to the hosts of SCMV: sugarcane and maize, mainly.
According to Chaves and Ortiz (2012), Mexican isolate JAL-1, with access number GU474635 has a higher phylogenetically proximity to SCMV isolates from Brazil and the United States, while CP sequences reported in China and Germany are phylogenetically nearer to the Mexican isolation SCMV-VER1, with access number EU091075. In this study, the Mexican isolates mentioned above were closely related and they grouped with sequences from China and Germany, composing group 1. On the other hand, Moradi et al. (2017), also contemplated in their analysis the same two Mexican sequences of SCMV and placed them in two different groups. Due to the above, there may be a need to include more HC-Pro sequences of SCMV into the phlogenetic analysis to observe a similar tendency to those reported by these authors.
Conclusions
The RT-PCR tests carried out on symptomatic and asymptomatic vegetative sugarcane material helped identify the presence of SCMV in the states of Colima, Jalisco, and Nayarit. This virus is widely distributed in the sugarcane areas of western Mexico, probably due to the presence of vector aphids and the propagation of potentially infected stake seed. The phylogenetic analysis using partial HC-Pro sequences from the SCMV isolate JalMex-126 helped identify two groups, the first of which had maize plants as host, and the second, with host plants of different species and sugarcane hybrids. Isolate JalMex-126 of this study may probably share common genetic origins with isolates from India, Australia, China, and Argentina.
Literatura citada
Balarabe DD, Adama Y, Azmat KUU, Aisha ZM. 2014. Identification of virus isolates inducing mosaic of sugarcane in Makarfi local government area of Kaduna state, Nigeria. African Journal of Biotechnology 13:1351-1357. http://dx.doi.org/10.5897/AJB2013.13467 [ Links ]
Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L, Zurcher EJ (eds). 1996. `Plant Viruses Online: Descriptions and Lists from the VIDE Database. Version: 16th January 1997. Disponible en línea: http://biology.anu.edu.au/Groups/MES/vide/ (Consulta, julio de 2017). [ Links ]
Chaves BG, Espejel F, Alcalá BRI, Hernández VJ, Silva RS. 2011. Short distance movement of genomic negative strands in a host and nonhost for Sugarcane mosaic virus (SCMV). Virology Journal 8:15. https://doi.org/10.1186/1743-422X-8-15 [ Links ]
Chaves BG, Ortiz RLY. 2012. Evidencia de orígenes filogenéticos diferentes de dos aislamientos mexicanos del virus del mosaico de la caña de azúcar (SCMV). Acta Agronómica 61:79-87. Disponible en línea: https://revistas.unal.edu.co/index.php/acta_agronomica/article/view/32464 [ Links ]
CONADESUCA, Comité Nacional para el Desarrollo Sustentable de la Caña de Azúcar. 2015. Fitopatologías. http://www.conadesuca.gob.mx (Consulta, junio de 2016). [ Links ]
Dallwitz MJ 1980. A general system for coding taxonomic descriptions. Taxon 29:41-46. http://delta-intkey.com/www/dallwitz-1980.htm [ Links ]
Dallwitz MJ, Paine TA, Zurcher EJ. 1993. User’s Guide to the DELTA System: a general system for processing taxonomic descriptions. http://delta-intkey.com/www/uguide.htm [ Links ]
Engebrecht J, Brent R, Kaderbhai MA. 2001. Minipreps of Plasmid DNA. Current Protocols in Molecular Biology. 15:II:1.6:1.6.1-1.6.10. DOI: 10.1002/0471142727.mb0106s15 [ Links ]
Espejel F, Jeffers D, Noa CJC, Ruiz CS, Silva RL. 2006. Coat protein gene sequence of a Mexican isolate of Sugarcane mosaic virus and its infectivity in maize and sugarcane plants. Archives of Virology 151:409-412. DOI: 10.1007/s00705-005-0645-3 [ Links ]
Figueredo L, Hernández L, Linares B. 2004. Relación epidemiológica entre áfidos (Homoptera: Aphididae) y enfermedades virales en el cultivo caña de azúcar en los valles de los ríos Turbio y Yaracuy, Venezuela. Caña de azúcar 22:5-19. [ Links ]
Filippone MP, Perera MF, Salgado M, García MG, Vellicce GR y Castagnaro P. 2010. Diagnóstico molecular de enfermedades sistémicas de la caña de azúcar en la Argentina: ajuste metodológico y aplicaciones. Revista industrial y agrícola de Tucumán 87:1-11. Disponible en línea: http://www.scielo.org.ar/scielo.php?script=sci_abstract&pid=S1851-30182010000200001 [ Links ]
Gonçalves MC, Pinto LR, Souza SC and Landell MGA. 2012. Virus diseases of sugarcane. A constant challenge to sugarcane breeding in Brazil. Functional Plant Science and Biotechnology 6:108-116. Disponible en línea: http://www.globalsciencebooks.info/Online/GSBOnline/images/2012/FPSB_6(SI2)/FPSB_6(SI2)108-116o.pdf [ Links ]
Gonçalves MC, Santos AS, Maia IG, Chagas CM and Harakava R. 2007. Characterization of an isolate of Sugarcane mosaic virus breaking down resistance of commercial sugarcane varieties. Fitopatologia Brasileira 32:32-39. http://dx.doi.org/10.1590/S0100-41582007000100004 [ Links ]
Grisham MP and Pan YB. 2007. A genetic shift in the virus strains that cause mosaic in Louisiana sugarcane. Plant Disease 91: 453-458. https://doi.org/10.1094/PDIS-91-4-0453 [ Links ]
Grisham MP. 1994. Strains of Sorghum mosaic virus causing sugarcane mosaic in Louisiana. Plant Disease 78:729-732. Disponible en línea: https://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1994Articles/PlantDisease78n07_729.pdf [ Links ]
Grisham MP. 2000. Mosaic. Pp 249-254. In: P Rott, RA Bailey, JC Comstock, BJ Croft, AS Saumtally (eds). A Guide to Sugarcane Diseases. CIRAD Publications Service. Montpellier, France. 339p. [ Links ]
Ha C, Revill P, Harding RM, Vu M and Dale JL. 2008. Identification and sequence analysis of potyviruses infecting crops in Vietnam. Archives of Virology 153:45-60. http://dx.doi.org/10.1007/s00705-007-1067-1 [ Links ]
Hall JS, Adams B, Parsons TJ, French R, Lane LC and Jensen SG. 1998. Molecular cloning, sequencing and phylogenetic relationships of a new potyvirus: Sugarcane streak mosaic virus and a reevaluation of the classification of the Potyviridae. Molecular Phylogenetics and Evolution 10:323-332. https://doi.org/10.1006/mpev.1998.0535 [ Links ]
Hema M, Joseph J, Gopinath K, Sreenivasulu P and Savithri HS. 1999. Molecular characterization and interviral relationships of a flexuous filamentous virus causing mosaic disease of sugarcane (Saccharum officinarum L.) in India. Archives of Virology 144:479-490. Disponible en línea: http://studylib.net/doc/13774806/molecular-characterization-and-interviral-relationships [ Links ]
Koike H, Guillaspie AG. 1989. Mosaic. Pp 287-288. In: C Ricaud, BT Egan, AG Gillaspie Jr, CG Hughes (eds). Disease of sugarcane. Major Disease. Elsevier. Amsterdam, The Netherlands. 387p. [ Links ]
Moradi Z, Nazifi E and Mehrvar M. 2017. Occurrence and evolutionary analysis of coat protein gene sequences of Iranian isolates of Sugarcane mosaic virus. The Plant Pathology Journal 33:296-306. DOI: 10.5423/PPJ.OA.10.2016.0219 [ Links ]
Raid RN, Comstock JC. 2000. Common rust. Pp 85-89. In: P Rott, RA Bailey, JC Comstock, BJ Croft, AS Saumtally (eds). A Guide to Sugarcane Diseases. CIRAD Publications Service. Montpellier, France. 339p. [ Links ]
Rao GP, Chatenet M, Girard JG and Rott P. 2006. Distribution of sugarcane mosaic and Sugarcane streak mosaic virus in India. Sugar Technology 8:79-81. https://doi.org/10.1007/BF02943747 [ Links ]
Riley SP, Woodman ME and Stevenson B. 2008. Culture of Escherichia coli and Related Bacteria. Current Protocols Essential Laboratory Techniques. 00:4.2:4.2.1-4.2.25. DOI: 10.1002/9780470089941.et0402s00 [ Links ]
Saumtally AS and Sullivan S. 2000. Brown spot. Pp 77-80. In: P Rott, RA Bailey, JC Comstock, BJ Croft, AS Saumtally (eds). A Guide to Sugarcane Diseases. CIRAD Publications Service. Montpellier, France. 339p. [ Links ]
Sawazaki HE, Nogueira SLA, Gonçalves CRNCB, Arruda VRF and Colombo CA. 2013. Molecular diagnosis optimization of virus, bacteria and fungi in sugarcane. International Research Journal of Plant Science 4:76-83. Disponible en línea: http://www.interesjournals.org/IRJPS [ Links ]
Singh M, Singh A, Upadhyaya PP and Rao GP. 2005. Transmission studies on an Indian isolate of sugarcane mo-saic potyvirus. Sugar Technology 7:32-38. https://doi.org/10.1007/BF02942526 [ Links ]
Singh V, Sinha OK and Kumar R. 2003. Progressive decline in yield and quality of sugarcane due to sugarcane mosaic virus. Indian Phytopathology 56:500-502. [ Links ]
Smith GR and Van de Velde R. 1994. Detection of Sugarcane mosaic virus and Fiji disease virus in diseased sugarcane using the polymerase chain reaction. Plant Disease 78:557-561. Disponible en línea: http://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1994Articles/PlantDisease78n06_557.pdf [ Links ]
Viswanathan R, Balamuralikrishnan M and Karuppaiah R. 2008. Characterization and genetic diversity of Sugarcane streak mosaic virus causing mosaic in sugarcane. Virus Genes 36(3):553-564. DOI:10.1007/s11262-008-0228 [ Links ]
Whittle PJL and Irawan. 2000. Pokkah boeng. Pp 136-140. In: P Rott, RA Bailey, JC Comstock, BJ Croft, AS Saumtally (eds). A Guide to Sugarcane Diseases. CIRAD Publications Service. Montpellier, France. 339p. [ Links ]
Xie J, Wang M, Xu D, Li R and Zhou G. 2009. Simultaneous detection and identification of four sugarcane viruses by one-step RT-PCR. Journal of virological methods 162:64-68. Disponible en línea: https://docslide.net/documents/simultaneous-detection-and-identification-of-four-sugarcane-viruses-by-one-step.html [ Links ]
Xie X, Chen W, Fu Q, Zhang P, An T, Cui A, An D. 2016. Molecular Variability and Distribution of Sugarcane Mosaic Virus in Shanxi, China. PLoS ONE 11: e0151549. https://doi.org/10.1371/journal.pone.0151549 [ Links ]
Xu DL, Park JW, Mirkov TE and Zhou GH. 2008. Viruses causing mosaic disease in sugarcane and their genetic diversity in southern China. Archives of Virology 153:1031-1039. https://doi.org/10.1007/s00705-008-0072-3 [ Links ]
Yang ZN and Mirkov TE. 1997. Sequence and relationships of sugarcane mosaic and Sorghum mosaic virus strains and development of RT-PCR-based RFLPs for strain discrimination. Phytopathology 87(9):932-939. Disponible en línea: https://www.ncbi.nlm.nih.gov/pubmed/18945064 [ Links ]
Zhang YL, Pennerman KK, Wang H and Yin G. 2016. Characterization of a Sorghum mosaic virus (SrMV) isolate in China. Saudi Journal of Biological Sciences 23:237-242. https://doi.org/10.1016/j.sjbs.2015.02.013 [ Links ]
Received: September 11, 2017; Accepted: October 16, 2017











 text in
text in