Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.35 no.3 Texcoco sep. 2017
https://doi.org/10.18781/r.mex.fit.1703-4
Review articles
Vasconcellea cauliflora resistance to Papaya ringspot potyvirus (PRSV-P) and its introgression in Carica papaya
1Instituto de Biociencias, Universidad Autónoma de Chiapas, Blvd. Príncipe Akishino s/n colonia Solidaridad 2000, Tapachula, Chiapas, C.P. 30798, México.
2Campo Experimental Bajío-INIFAP, Km 6.5 carretera Celaya a San Miguel de Allende, Celaya, Guanajuato, C.P. 38010, México.
3Universidad de Guanajuato, División de Ciencias de la Vida, Km 9 carretera Irapuato-Silao, ExHda. El Copal, Irapuato, Guanajuato, C.P. 36500, México.
4Campo Experimental Valle de México-INIFAP, Km. 13.5 carretera Los Reyes-Texcoco, Coatlinchán, Texcoco Estado de México, C.P. 56250, México.
Papaya is a crop of major economic importance, but it is threatened by the presence of viral diseases that are mainly transmitted by vector insects. Although conventional pest control practices seem to keep out the spread of the virus, it is not enough to avoid production loses. There are some molecular tools from genetic engineering to obtain disease resistant plants with potential to solve the problem. This review is an information compilation about the problem caused by virus with more impact on papaya production: Papaya ringspot potyvirus (PRSV-P) and Papaya mosaic virus (PapMV), their genetic variability and some strategies used to develop disease resistant hybrids resulting from the recombination between Vasconcellea genus and Carica papaya commercial varieties.
Key words: Intergeneric hybrids; viruses; genetic improvement
El cultivo de papaya (Carica papaya L.) posee gran importancia económica, la cual es afectada por enfermedades causadas por virus, principalmente transmitidas por áfidos vectores. Aunque las prácticas convencionales de control de vectores parecen mantener al margen la propagación del virus, no es suficiente para evitar pérdidas en la producción. Existen desarrollos de la ingeniería genética que permiten obtener plantas resistentes a las enfermedades virales. En la presente revisión, se aborda la problemática asociada a los virus con mayor impacto para el cultivo de la papaya: Papaya ringspot potyvirus (PRSV-P) y el Papaya mosaic virus (PapMV), su variabilidad genética y las estrategias de mejoramiento genético que han permitido la generación de poblaciones resistentes a través de la recombinación de especies del género Vasconcellea con variedades comerciales de Carica papaya.
Palabras clave: Híbridos intergenéricos; virus; mejoramiento genético
The family Caricaceae is classified in six groups formed by 35 species widely distributed worldwide. Two species of the Cylicomorpha genus are in tropical Africa, while the endemic Horovitzia and Jarilla genera prevail in Central America, and Jarilla and Vasconcellea, in South America (Antunes and Renner, 2014). Carica papaya wild species are widely distributed in tropical forests from Mexico up to Costa Rica (Chávez-Pesqueira et al., 2014). The hermaphrodite version of C. papaya currently sown is the result of domestication by Mexican ethnic groups, presumably the Mayas (VanBuren et al., 2015).
For this reason, Central and South American countries are considered the centers of genetic and biological diversity and suitable for papaya commercial production. However, in Mexico, papaya production is affected by major relevant phytopathological problems such as viral diseases caused by vector insects resulting in losses of 90% in plantations, i.e., reductions in plant density (Mora et al., 1992).
At least 12 viruses are reported worldwide that pose a threat to papaya production: Papaya ringspot virus (PRSV-P), Papaya leaf distortion mosaic virus (PLDMV), Zuchinni yellow mosaic virus (ZYMV), Papaya mosaic virus (PapMV), Papaya leaf curl virus (PaLCV), Chilli leaf curl virus (ChiLCuV), Tomato leaf curl New Delhi virus (ToLCNDV), Croton yellow vein mosaic virus (CYVMV), Papaya droopy necrosis virus (PDNV), Tomato spotted wilt virus (TSWV), Papaya lethal yellowing virus (PLYV) and Papaya meleira virus (PMeV) (Mishra et al., 2015).
From these viruses, those of major importance in Mexico are PRSV-P, PapMV and PMeV, which are a problem because of the economic losses they cause (Silva-Rosales et al., 2010; Rivas-Valencia et al., 2008). Infections produced by those viruses reduce fruit yield per hectare that in turn reduces up to 275% the net income per hectare (Yorobe, 2009). The highest impact on papaya production is mainly due to the fact that none of the commercial varieties has genetic resistance to PRSV-P or PMeV (Abreu et al., 2014; Singh et al., 2005). However, some non-commercial varieties have been reported to have low levels of susceptibility (Rodríguez et al., 2013).
PRSV-P is one of the viruses causing the greatest damage to papaya productivity with values higher than 70% (Tennant et al., 2007); in contrast, the effects of PapMV are not so aggressive. Papaya naturally hosts both virus at the same time (Noa-Carranza et al., 2006), and for this reason two events may take place: 1) Synergism, which is the primary PRSV-P infection followed by PapMV infection; 2) Antagonism, that results from the first PapMV infection followed by PRSV-P infection, which suggests that PapMV activates a cascade pathway in defense response in the infected plant (Chávez-Calvillo et al., 2016).
An alternative for developing genetic resistance to PRSV-P is to genetically transform somatic embryos obtained in vitro (or any other explant) using Agrobacterium (strain C58-Z707) or particle pumping. The gen transferred to papaya is the one encoding the capsid protein of a specific virus isolate. However, if the transgene is built with the sequence of a local virus, there is a relatively high probability that the transgenic plants do not develop resistance to viruses from other geographical origin (Mishra et al., 2015). Two transgenic varieties of papaya of the Solo morphotype are currently available in the international market (Rainbow and SunUp) (Mishra et al., 2015) that are characterized by being small fruits weighing from 300 to 600 g; they were modified using the PRSV-P HA 5-1 strain originally from Hawaii (Gonsalves, 2006).
As an additional alternative, 31 varieties of C. papaya (primary genetic pool) were evaluated to find genetic resistance, but only tolerance was found in the Califlora variety (Roff, 2007). For this reason, genetic resistance to PRSV-P was identified in Vasconcellea cauliflora species (secondary genetic pool) that was later introduced (introgression) in C. papaya materials using conventional improvement techniques (Yanthan et al., 2017).
Given that in Mexico the establishment of transgenic papaya is banned (NOM-056-FITO-1995), this review is intended to update progress on introgression of resistance to the PRSV-P, virus from some species of the Vasconcellea genus by recombining them with C. papaya, as an alternative for developing genetic resistance.
Viral genetic diversity
PRSV belongs to the genus Potyvirus, family Potyviridae. These viruses are transmitted by seed, mechanical means and over 25 aphid species in a non-persistent way, a distinctive feature of the group. There are two biotypes: “P” that infects papaya and “W” that damages some members of the cucurbit family, such as watermelon (Citrullus lanatus) and melon varieties (Cucumis melon) (Tripathi et al., 2008).
The genetic variability of PRSV-P is as wide as the diversity of ecological niches where papaya is grown. According to Bateson et al. (2002), there is 12% of variability in the nucleotidic sequence among geographic regions such as Asia, Australia and North America. It has also been suggested that it first appeared in Asia from where it spread to the rest of the world.
In Mexico, PRSV-P was first reported in the state of Veracruz in 1964. So far, five genetic groups have been reported to be distributed across the Pacific coast, the Gulf coast, Cotaxtla regions (Veracruz), the Yucatán peninsula and the western Pacific coast (Noa-Carranza et al., 2006). Within this diversity, two isolates from Veracruz and one from Chiapas are highly correlated with isolates from North America and Australia, they are also genetically different from those from China, Taiwan, Vietnam and India (Silva-Rosales et al., 2000).
Twenty-two complete PRSV-P genomes from several countries have been sequenced, and the phylogenetic analyses show that the isolates are grouped according to their country of origin. Therefore, Taiwan, China, Thailand and India form one group, and a second group includes isolates from the Americas (Brazil, Mexico, United States) (Mishra et al., 2015). In contrast, nucleotide analyzes of the capsid protein genes from virus samples collected in Cuba (7 samples) and Brazil (21 samples) show that the genetic distance among them ranged from 0% to 9.2%. Analyses performed in each country showed formation of genetic subgroups according to the geographic regions where the samples were taken, i.e., there is higher similarity among isolates from neighboring regions. However, when genetic information from the virus GenBank was included, two groups with 86% maximum genetic similarity were identified. The first group included viruses from India, Hawaii, Mexico, Venezuela, Cuba and Brazil, and the second, viruses from China, Taiwan, Thailand, South Korea and Malaysia (Rodríguez et al., 2014).
The Papaya Mosaic virus (PapMV) is a member of the Potexvirus genus and the family Alfaflexiviridae (Mishra et al., 2016). It was first reported in Mexico in 2001 (Noa-Carranza et al., 2006). Comparisons of protein coat of strains from Canada with strains from China showed 98% similarity (Sit et al., 1989). However, comparisons between strains from China and strains from Mexico showed 73% similarity (Tennant et al., 2007), whereas in Mexico strains of that virus showed similarity higher than 91% (Noa-Carranza et al., 2006).
In general, it seems that the genetic variability of viruses PRSV-P and PapMV makes it necessary to use isolates from each region for genetic transformation events, to create attenuated viruses or develop conventional schemes for selecting papaya genetic materials for virus tolerance. Both are present in diverse ecological regions of Mexico (Figure 1).
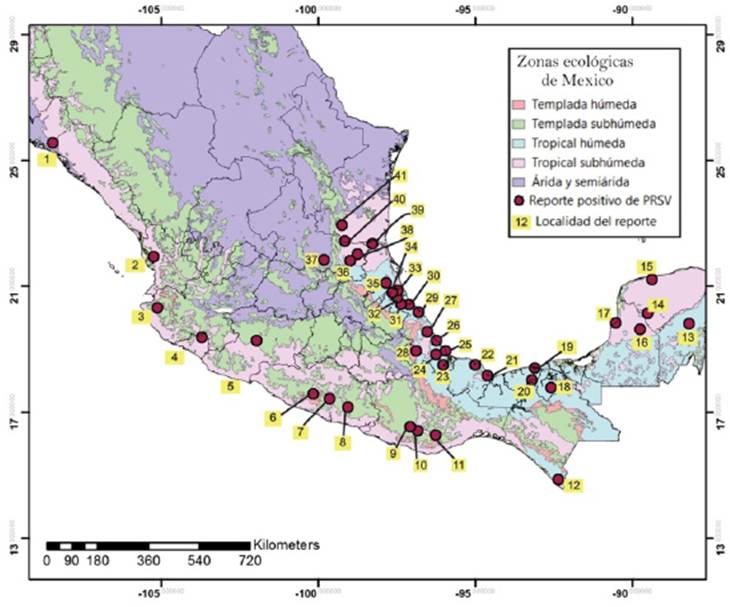
Figure 1 Distribution of PRSV and PapMV in Mexico. Developed by the authors based on a study on virus-infected plants conducted by Noa-Carranza et al. (2006) in 41 locations.
Symptoms caused by PRSV-P AND PapMV
Characteristic symptoms of PRSV-P virus-infected plants include mosaic, leaf deformation, yellowing or chlorosis (Figure 2), stunting, ring spots on fruit, mosaic spots, and oil and water-soaked bands on petioles and the upper part of the stem. Infected plants develop abnormally, are smaller and have low vigor and less production and commercial quality (Kumar et al., 2014). Changes observed in mass accumulation in PRSV-P virus infected plants are associated with a decrease in the photosynthesis rate and an increase in the respiration rate (Marler et al., 1993).
Symptoms of PapMV infected plants include mottling of young leaves, chlorosis of veins and leaf lamina curved towards the underside (Figure 2); also, infected plants are shorter and have small leaves (Taylor, 2001). However, a co-infection by both viruses produces apical leaf necrosis, twisting, mottling and mosaic (Noa-Carranza et al., 2006).

Figure 2 Symptoms produced by PRSV-P and PapMV in a leaf of Carica papaya L. Maradol variety. A) Leaf showing symptoms of PRSV-P at intermediate phase. B) Leaf showing PRSV symptoms at advanced phase. C) Leaf showing symptoms of PapMV at intermediate phase. D) Leaf showing symptoms of PapMV at advanced phase. Tapachula, Chiapas, Mexico, 2016.
The presence and severity of symptoms produced by both viruses depend on diverse environmental factors that, when present, can increase or mitigate the severity of the infection. The susceptibility of plantlets inoculated with PRSV-P increases when they are subjected to short periods of darkness because the carbohydrate content in leaves decreases, which may interfere with the establishment of the virus in the cells. Temperature is also crucial for disease development. According to Cabrera et al. (2010), when temperatures are higher than 40 °C, the viral accumulation decreases; the same occurs when temperatures are lower than 20 °C. However, when room temperature ranges between 26 °C and 31 °C, the viral accumulation and expression of symptoms reaches its maximum level. This could be because the defense mechanism of silencing regulated by ARN can be easily suppressed by the environmental temperature (Mangrauthia et al., 2009).
Strategies for managing viral infections
Management strategies for diseases associated with virus infections include using virus-free propagules, removing plants with early infections, establishment of planting dates, control and study of vector aphid populations, transmission interference by aphids (plant barriers), mineral oil application, cross protection, genetic improvement for resistance, development of transgenic plants and quarantine practices (Villegas, 2001; Rivas-Valencia et al., 2003; 2008; Subramanya and Zitter, 2014).
The most common strategy for controlling vector aphids is insecticide spraying. However, it seems possible to develop variants resistant to specific molecules such as neonicotinoids (Koo et al., 2014). For this reason, the populations dynamics is important in disease incidence analyses in papaya plantations. For example, Mora-Aguilera et al. (1996) state that the transplanting location and date (in February, April or June) are important agronomic factors to make them coincide with a lower incidence of infected plants in the central region of the state of Veracruz, Mexico.
Cross protection involves the use of attenuated virus to infect the plant and activate its genic silencing mechanism before it becomes infected by virulent strains (Fermin et al., 2010). In Taiwan, Thailand and Hawaii, two attenuated isolates have been used in papaya production: PRSV-P HA 5-1 and PRSV-P HA 6-1. Although, these isolates do not provide total protection, papaya growers obtain acceptable yields consistently and predictably (Subramanya and Zitter, 2014).
Development of transgenic papaya
There are tree strategies to produce papaya resistant to PRSV-P (Villegas, 2001): 1) interspecific crosses aimed at developing a moderate-tolerant variety to be used as a genetic source; 2) interspecific crosses between C. papaya and its resistant wild relatives (such as Vasconcellea cauliflora, V. quercifolia and V. pubescens) to develop resistant hybrids; and 3) using genetic engineering to obtain resistance by transferring the gene of the virus capsid to papaya grown and develop transgenic papaya.
In 1981, Hawaii’s papaya improvement program implemented the use of biotechnological tools such as: in vitro tissue culture for micropropagation of lines, rescue of embryos of interspecific crosses and genetic engineering (Villegas, 2001). Papaya was conceived to be transgenic by inserting the gene that encodes the protein coat of the PRSV virus HA 5-1 strain in the papaya genome. Later, tests were conducted in the field using mechanical inoculation to induce infection with aphids, finally 95% of the evaluated individuals showed resistance. However, the transgene was only efficient in protecting papaya against the local virus but not against PRSV from Taiwan, Thailand and Mexico (Gonsalves, 2006). These results confirmed that transgenesis requires a compatibility level higher than 98% between the virus and the transgene, and that the diversity of PRSV-P strains in the world’s ecological niches limits their use (Gonsalves, 1998).
On the other hand, Davis and Ying (2004) transformed papaya lines using genes of the protein coat of PRSV-P H1K isolate from Florida. It should be noted that, after transformation, the percentage of susceptible plants was associated with the molecular architecture of the transgene used for transformation, the number of copies of the transgene present in the tissue and the effect of the genetic background on the varieties crossed with transgenic lines.
Currently, in Hawaii, over 70% of the area devoted to papaya cultivation corresponds to the transgenic hybrids SunUp and Rainbow (Azad et al., 2014). Due to this tendency, countries like Jamaica, Venezuela and Thailand created their own technology to develop genetic materials resistant to PRSV-P endemic strains. In recent years, transgenic papaya has been established in Australia, Jamaica, Venezuela, Vietnam, Thailand, Taiwan and the Philippines (Fermin et al., 2010).
In Mexico, there are legal restrictions for the establishment of commercial transgenic papaya plantations, as stated in the Norma Oficial Mexicana NOM-056-FITO-1995. Between 1995 and 2015, COFEPRIS evaluated food safety and authorized 146 events of genetically modified products but did not include papaya (COFEPRIS, 2015). This is a measure to protect plant centers of origin and domestication, as is the case of papaya in Mexico.
Introgression of resistance genes in virus
Gene introgression among species occurs naturally and has enhanced genetic diversity through cross-pollination (Droogenbroeck et al., 2006). This also allows the development of new varieties because disease resistance genes from wild species can be introgressed into important commercial varieties (Dinesh et al., 2013).
Interspecific hybridization is the result of a long-term genetic strategy for controlling viral infections that consists in making artificial crosses between domesticated papaya (C. papaya L.) and other species of the Vasconcellea genus that is a member of the family Caricaceae (Drew, 2014).
There are 21 Vasconcellea species (Coppens d’Eeckenbrugge et al., 2014) in Central and South America, and this will allow us to evaluate genetic material with potential resistance to native viruses that could be used to develop resistant populations. However, in this process, it is important to consider both the presence of prezygotic and postzygotic barriers to produce useful hybrid seeds (poor pollen germination and sterile seed development, due to embryo and ovule abortion) (Veena and Dinesh, 2013), and the genetic distances among species in crossing schemes (Sharma and Tripathi, 2016). For this reason, introgression requires identifying species and varieties that are resistant to PRSV-P and also compatible in artificial crosses, to serve as parents.
In Mexico, the importance of evaluating wild relatives of C. papaya as sources of genetic resistance to PRSV-P and PapMV was recognized since 1978. At that time, Vasconcellea cauliflora (Vc), V. pubescens, V. stipulata and V. candicans were reported as being resistant to papaya ringspot virus (PRSV) and Vc is the only one found in Mexico. However, crosses between C. papaya and Vasconcellea spp. exhibited compatibility barriers, such as development of F1 seed without embryo and F1 seed without endosperm (Mosqueda, 1978). When parents show low compatibility, a protocol is needed to rescue immature hybrid embryos using in vitro tissue culture techniques (Azad et al., 2014).
Some Vasconcellea species are resistant to certain PRSV-P strains; for example, V. quercifolia is resistant to virus strains from Florida, Hawaii and Australia but susceptible to isolates from Venezuela; V. cauliflora is considered to be resistant to strains from Maracay (Venezuela), Mexico, Australia and India but susceptible to isolates from Florida and some from Venezuela; other species are resistant: V. pubescenes, V. stipulata, V. heilbornii and V. candicans (Horovitz and Jimenez, 1967; Sharma and Tripathi, 2016). Results from these experiments showed resistance variability in terms of the geographic origin of PRSV-P.
Horovitz and Jimenez (1967) made interspecific crosses and found some hybrids resistant to PRSV-P. The authors also stated that a dominant gene is responsible for resistance to virus infections. In crosses between papaya and V. quercifolia several fertile hybrids have been obtained (Villegas, 2001). In 2011, segregant populations were obtained from four backcrosses (Backcross: BC1 to BC4) between V. quercifolia and C. papaya lines. After mechanical inoculation with PRSV-P isolated in the Philippines, plantlets without symptoms (72.3% to 96.1%) under greenhouse conditions were recorded. After 18 months in the field, plants from the BC4 backcross were analyzed using the ELISA technique but only 66% of them were resistant (Siar et al., 2011). In BC3 plants, the ratio of resistant: susceptible segregants was 3:1, whereas in BC4 plants two types of ratio were observed: 2:1 and 4:1. These results indicated that the resistance to virus infection is polygenic (Alamery and Drew, 2014). Although in combinations of C. papaya x V. pubescens resistance to PRSV-P was also found, their progeny was sterile to continue a conventional improvement scheme. For this reason, a bridge has been proposed to transfer resistance genes through V. pubescens x V. parviflora crosses, and then make crosses with C. papaya (Drew, 2014). On the other hand, Kumar and Tripathi (2016) proposed another transference bridge by intercrossing V. cundinamarcensis x V. parviflora, and then pollinate F1 individuals with C. papaya to obtain hybrids resistant to virus infections.
Genetic studies using an F2 generation from V. pubescens (resistant) x V. parviflora (susceptible) crosses showed a STK (serine threonine protein kinase marker gene) associated with resistance to PRSV-P. Based on this result, Haireen and Drew (2014) cloned the STK gene and observed that its genomic sequence is similar to that of Carica papaya (known as: CP_STK). They also found an equivalent gene in V. pubesces (VP_STK2). When they compared their genomic sequences, they noted structural differences and, for this reason, the protein products may show a differentiated role, a fact that may influence the species susceptibility or resistance to the virus.
Intergeneric hybrids with Vasconcellea cauliflora
Magdalita et al. (1997) made C. papaya x Vc crosses and rescued in vitro F1 embryos to develop hybrid individuals. The hybrid plants were artificially inoculated with two PRSV-P isolates from Australia. From 114 plants inoculated in the greenhouse, 22 survived and 100% did not develop virus infection symptoms. In another lot of 20 hybrid plants established in the open field, 12 survived and none of them developed virus infection, a fact that was confirmed using the ELISA technique.
There are currently protocols for in vitro tissue culture to rescue embryos resulting from manual pollination of papaya with Vc pollen. It has been stated that the optimum date to extract embryos from immature seeds is 90 days after pollination (Azad et al., 2014; Vegas et al., 2003).
Another alternative for obtaining intergeneric embryos would be to break the compatibility barriers present in artificial crosses between papaya and Vc. For this purpose, Dinesh et al. (2007) applied a 5% sucrose solution to flower stigmas before manual pollination and obtained hybrid seeds in crosses between Surya and Pusa Dwarf papaya varieties x Vc. 71.1% of pollinated flowers bore fruit and produced 13.7 F1 seeds in average. Using ISSR molecular markers, the authors confirmed that the seeds were interspecific hybrids. Later, Dinesh et al. (2013) created a selection scheme making crosses between C. papaya var. Arka Surya x Vc (Figure 3) and achieved encouraging results (Yanthan et al., 2017).
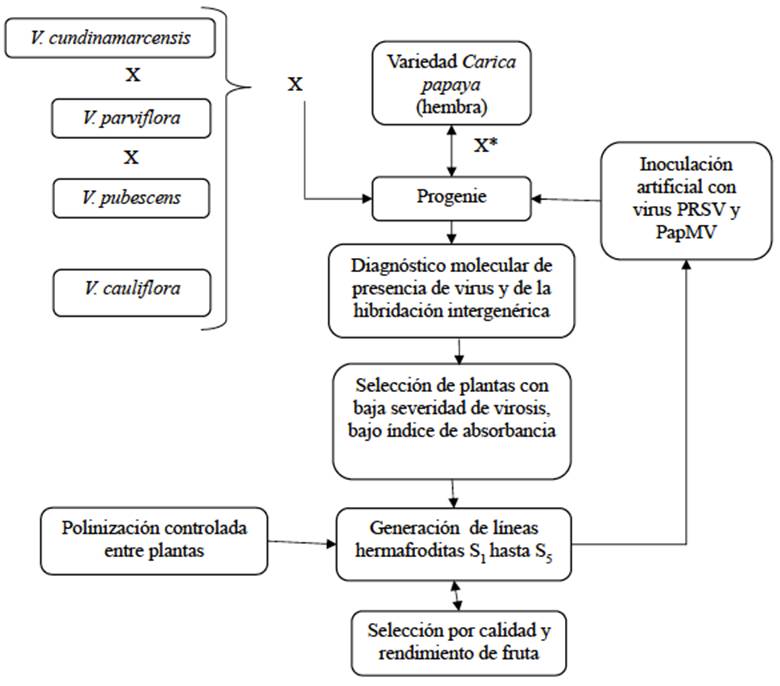
Figure 3 Scheme of crosses between Vasconcellea and Carica papaya species to develop lines tolerant to virus infections. It is important to use DNA markers to confirm intergeneric hybrids, and ELISA tests to estimate the level of viral charge in the tissue of the plants. Based on Dinesh et al. (2013) and Kumar and Tripathi (2016). X= Cross, X*= Backcross.
Jayavalli et al. (2011, 2012) made crosses between Vc and 9 papaya varieties from India to which they also applied 5% sucrose solutions combined with calcium chloride and boron. From the total of pollinated flowers (n=1197), 25.7% bore fruits. Each fruit had 2.3 hybrid seeds in average, and from the total number of seeds, 58.1% germinated. F1 plants performed well and were inoculated with PRSV-P; 23.6% did not develop viral symptoms, but the remaining percentage exhibited mosaic and deformation of merismatic leaves, so it was determined that Pusa Nanha x Vc was the best intergeneric hybrid.
Sudha et al. (2013) evaluated an F2 generation of crosses between Vc and Pusa Nanha, CP50 and CO7 varieties. From 700 F2 plants inoculated with PRSV-P, 46% of them did not show virus infection symptoms. Considering the number of days from the inoculation date to the appearance of symptoms, as well as their severity, the best F2 family resulted from the cross between Pusa Nanha x Vc. Using an F3 generation, Sudha et al. (2015) evaluated 1,778 F3 plants inoculated with PRSV-P and found that 30.7% did not show virus infection symptoms after 27 days; therefore, according to the severity scale, they were considered resistant; the remaining percentage of plants were classified as susceptible and highly susceptible. When the resistant plants were taken to the greenhouse and evaluated up to 270 after inoculation, plants from the cross between Pusa Nanha x Vc were found to be resistant- and intermediate resistant.
Jayavalli et al. (2015) also studied an F2 generation from crosses CO7 x Vc, Pusa Nanha x Vc y CP50 x Vc. For this purpose, they inoculated 683 plantlets of the three crosses with PRSV-P, and found that 12.8% did not show infection symptoms, but in a virus detection trial using DAS-ELISA they obtained low absorbance values (< 0.24). In a CO7 x Vc cross, they obtained resistant plants in a 15:1 ratio (susceptible:resistant), while in the other two crosses (Pusa Nanha x Vc and CP50 x Vc), the resistant plants segregated in a 13:3 ratio.
The genetic structure proposed by Jayavalli et al. (2015), where they suggested that the presence of two dominant genes may explain such ratio of susceptible individuals, is shown in Table 1. They pointed out that there was no segregation in a 3:1 ratio as in the Mendelian inheritance. Also, the ratios observed indicate that there are multiple alleles in both species that when interacting produce genetic combinations associated with the PRSV-P virus.
Table 1 Genetic structure of C. papaya and V. cauliflora varieties and of their segregant populations in terms of their reaction to PRSV.
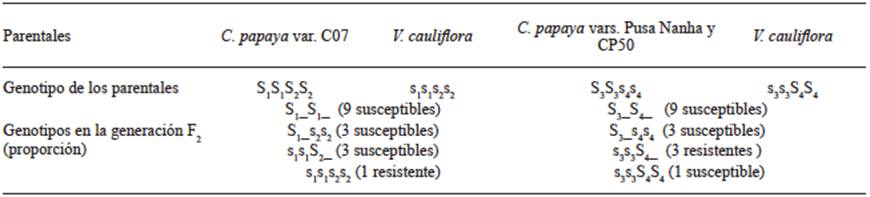
Source: Adapted from Jayavalli et al. (2015).
CONCLUSIONS
Studies on virus resistance have been mainly focused on PRSV-P due to its capacity to reduce papaya commercial yields. PapMV had not been fully addressed because of its low economic impact in papaya crops, but based on the evidence of co-infection and its effects on the severity of symptoms and damage to papaya crops, it is important to integrate PapMV in studies on virus resistance in Mexico.
Considering the importance of papaya crops and yield reductions attributable to PRSV-P and PapMV in Mexico, alternatives for developing varieties resistant to these viruses are required. Given the restrictions on using transgenic papaya varieties, an alternative is genetic improvement by making crosses between C. papaya lines and species of the Vasconcellea genus as donors for resistance to viruses, especially V. cauliflora that is naturally distributed across Mexico. It gives also an opportunity to include this kind of research in a genetic improvement program that can meet the demand of non-transgenic resistant varieties, through direct crosses or establishing a bridge to transfer virus-resistance genes using the interspecific triple cross among V. cundinamarcensis x V. parviflora x C. papaya to develop virus-resistant hybrids.
LITERATURA CITADA
Abreu PMV, Gaspar CG, Buss DS, Ventura JA, Ferreira PCG, and Fernández PMB. 2014. Carica papaya microRNAs are responsive to papaya meleira virus infection. PLoS ONE 9(7): e103401. https://doi.org/10.1371/journal.pone.0103401 [ Links ]
Alamery S and Drew R. 2014. Studies on the genetics of PRSV-P resistance genes in intergeneric hybrids between Carica papaya and Vasconcellea quercifolia. Acta Horticulturae 1022: 55-61. DOI: 10.17660/ActaHortic.2014.1022.5 [ Links ]
Antunes CF and Renner SS. 2014. The phylogeny of the caricaceae. Pp:81-92. In: R. Ming and Moore PH (eds.). Genetics and Genomics of Papaya. Springer Science Business Media, New York, USA. 438p. [ Links ]
Azad MAK, Amin L and Sidik NM. 2014. Gene technology for papaya ringspot virus disease management. The Scientific World Journal: 1-11. http://dx.doi.org/10.1155/2014/768038. [ Links ]
Bateson MF, Lines RE, Revill P, Chaleeprom W, Ha CV, Gibbs AJ and Dale JL. 2002. On the evolution and molecular epidemiology of the potyvirus papaya ringspot virus. Journal of General Virology 83:2575-2585. DOI: 10.1099/0022-1317-83-10-2575 [ Links ]
Cabrera MD, Cruz MM y Portal VO. 2010. Efecto de la temperatura en la virulencia del virus de la mancha anular de la papaya (PRSV-P). Fitosanidad 14(2):123-125. Disponible en línea: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S1562-30092010000200009 [ Links ]
Chávez-Calvillo G, Contreras-Paredes CA, Mora-Macías J, Noa-Carranza JC, Serrano-Rubio AA, Dinkova TD, Carrillo-Tripp M and Silva-Rosales L. 2016. Antagonism and synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya in determined by their order of infection. Virology 489:179-191. http://dx.doi.org/10.1016/j.virol.2015.11.026 [ Links ]
Chávez-Pesqueira M, Suárez-Montes P, Castillo G and Núñez-Farfán J. 2014. Habitat fragmentation threatens wild populations of Carica papaya (Caricaceae) in a lowland rainforest. American Journal of Botany 101(7):1092-1101. DOI: 10.3732/ajb.1400051 [ Links ]
COFEPRIS, Comisión Federal para la Protección contra Riesgos Sanitarios. 2015. http://www.cofepris.gob.mx/AZ/Paginas/OGMS/Cultivos.aspx . Consulta: Noviembre de 2016. [ Links ]
Coppens d’Eeckenbrugge G, Drew R, Kyndt T and Scheldeman X. 2014. Vasconcellea for Papaya Improvement. Pp:47-79. In: R Ming and Moore PH (eds.) Genetics and Genomics of Papaya. Springer Science Business Media, New York, USA . 438p. [ Links ]
Davis MJ and Ying Z. 2004. Development of papaya breeding lines with transgenic resistance to papaya ringspot virus. Plant Disease 88:352-358. https://doi.org/10.1094/PDIS.2004.88.4.352 [ Links ]
Dinesh MR, Rekha A, Ravishankar KV, Praveen KS and Santosh LC. 2007. Breaking the intergeneric crossing barrier in papaya using sucrose treatment. Scientia Horticulturae 114: 33-36. DOI: 10.1016/j.scienta.2007.05.010 [ Links ]
Dinesh MR, Veena GL, Vasugi C, Krishna MR and Ravishankara KV. 2013. Intergeneric hybridization in papaya for ‘PRSV’ tolerance. Scientia Horticulturae 161: 357-360. DOI: 10.1016/j.scienta.2013.07.009 [ Links ]
Drew R. 2014. The use of non-transgenic technologies for the development of papaya ringspot virus resistance in Carica papaya. Proc. Acta Horticulturae 1022:22-29. DOI: 10.17660/ActaHortic.2014.1022.2 [ Links ]
Droogenbroeck BV, Kyndt T, Romeijn-Peeters E, Thuyne WV, Goetghebeur P, Romero-Motochi JP and Gheysen G. 2006. Evidence of natural hybridization and introgression between Vasconcellea species (Caricaceae) from Southern Ecuador revealed by chloroplast, mithocondrial and nuclear DNA markers. Annals of Botany 97:793-805. DOI: 10.1093/aob/mcl038 [ Links ]
Fermin GA, Castro LT and Tennant PF. 2010. CP-Transgenic and non-transgenic approaches for the control of papaya ringspot: current situation and challenges. Transgenic Plant Journal 4 (Special Issue 1):1-15. Disponible en línea: http://www.globalsciencebooks.info/Online/GSBOnline/images/2010/TPJ_4(SI1)/TPJ_4(SI1)1-15o.pdf [ Links ]
Gonsalves D. 1998. Control of papaya ringspot virus in papaya: a case study. Annual Review of Phytopatology 36:415-437. https://doi.org/10.1146/annurev.phyto.36.1.415 [ Links ]
Gonsalves D. 2006. Transgenic papaya: development, release, impact and challenges. Advances in Virus Research 67:317-354. DOI: 10.1016/S0065-3527(06)67009-7 [ Links ]
Haireen MRR and Drew RA. 2014. Isolation and characterization of PRSV-P resistance genes in Carica and Vasconcellea. International Journal of Plant Genomics 5:145403. http://dx.doi.org/10.1155/2014/145403 [ Links ]
Horovitz S y Jiménez H. 1967. Cruzamientos interespecíficos e intergenéricos en caricáceas y sus implicaciones fitotécnicas. Agronomia Tropical 17(4): 323-343. [ Links ]
Jayavalli R, Balamohan TN, Manivannan N and Govindaraj M. 2011. Breaking the intergeneric hybridization barrier in Carica papaya and Vasconcellea cauliflora. Scientia Horticulturae 130:787-794. http://dx.doi.org/10.1016/j.scienta.2011.09.004 [ Links ]
Jayavalli R, Balamohan TN, Manivannan N and Robin S. 2012. Analysis of papaya intergeneric hybrids for morphological traits. Madras Agriculture Journal 99 (4-6): 166-170. Disponible en línea: https://www.cabdirect.org/cabdirect/abstract/20123213250 [ Links ]
Jayavalli R, Balamohan TN, Manivannan N, Rabindran R, Paramaguru P and Robin R. 2015. Transmission of resistance to papaya ringspot virus (PRSV) in intergeneric populations of Carica papaya and Vasconcellea cauliflora. Scientia Horticulturae 187:10-14. http://dx.doi.org/10.1016/j.scienta.2015.01.020 [ Links ]
Kassanis B. 2008. Some effects of high temperature on the susceptibility of plants to infection with viruses. Annals of Applied Biology 39:358-363. DOI: 10.1111/j.1744-7348.1952.tb01018.x [ Links ]
Koo HN, Jeong-Jin An, Sang-Eun P, Ju-Il K and Gil-Hah K. 2014. Regional susceptibilities to 12 insecticides of melon and cotton aphid, Aphis gossypii (Hemiptera: Aphididae) and a point mutation associated with imidacloprid resistance. Crop Protection 55:91-97. DOI: 10.1016/j.cropro.2013.09.010 [ Links ]
Kumar GR, Hohn T and Sharma P. 2014. Plant virus-host interaction. Molecular approaches and viral evolution. Pp:177-194. In: Gaur RK, Hohn T and Sharma P (eds.). Papaya Ringspot Virus-P: Overcoming Limitations of Resistance Breeding in Carica papaya L. Elsevier. USA. 408p. [ Links ]
Kumar SS and Tripathi S. 2016. Resistance against papaya ringspot virus in Vasconcellea species: present and potential uses. Pp:215-230. In: Gaur RK, Petrov NM, Patil BL and Stoyanova MI (eds.). Plant Viruses: Evolution and Management. Springer Science Business Media, Singapore. 312p. [ Links ]
Magdalita PM, Persley DM, Godwin ID, Drew RA and Adkins SW. 1997. Screening C. papaya x C. cauliflora hybrids for resistance to papaya ringspot virus-type P. Plant Pathology 46:837-841. DOI: 10.1046/j.1365-3059.1997.d01-90.x [ Links ]
Mangrauthia SK, Singh VPS, Jain RK, Praveen S. 2009. Ambient temperature perception in papaya for papaya ringspot virus interaction. Virus Genes 38:429-434. DOI: 10.1007/s11262-009-0336-3 [ Links ]
Marler TE, Mickelbart MV and Quituga R. 1993. Papaya ringspot virus influences net gas exchange of papaya leaves. HortScience 28(4): 322-324. Disponible en línea: http://hortsci.ashspublications.org/content/28/4/322.full.pdf [ Links ]
Mishra R, Kumar RG and Patil BL. 2015. Current knowledge of viruses infecting papaya and their transgenic management. Pp: 109-203. In: Gaur RK, Petrov NM, Patil BL and Stoyanova MI (eds.). Plant viruses: Evolution and Management. Springer Science Business Media, Singapore. 312 p. [ Links ]
Mora-Aguilera G, Nieto-Ángel D, Campbell CL, Téliz D and García E. 1996. Multivariate comparison of papaya ringspot epidemics. Phytopathology 86:70-78. DOI: 10.1094/Phyto-86-70 [ Links ]
Mosqueda VR. 1978. Papayo, Piña y Guayabo. Pp:299-309. In: Cervantes ST (ed.). Recursos Genéticos Disponibles a México. Sociedad Mexicana de Fitogenética A. C., Chapingo, México. 492p. [ Links ]
Noa-Carranza JC, González-de-León D, Ruiz-Castro BS, Piñero D and Silva-Rosales L. 2006. Distribution of papaya ringspot virus and papaya mosaic virus in papaya plants (Carica papaya) in Mexico. Plan Disease 90:1004-1011. http://dx.doi.org/10.1094/PD-90-1004 [ Links ]
Rivas-Valencia P, Mora-Aguilera G, Téliz-Ortiz D y Mora-Aguilera A. 2003. Influencia de variedades y densidades de plantación de papayo (Carica papaya L.) sobre las epidemias de mancha anular. Revista Mexicana de Fitopatología 21(2):109-116. Disponible en línea: http://www.redalyc.org/articulo.oa?id=61221203 [ Links ]
Rivas-Valencia P, Mora-Aguilera G, Téliz-Ortiz D y Mora-Aguilera A. 2008. Evaluación de barreras vegetales en el manejo integrado de la mancha anular del papayo en Michoacán, México. Summa Phytopathologica 34(4):307-312. http://dx.doi.org/10.1590/S0100-54052008000400001 [ Links ]
Rodríguez MD, Alonso M, Tornet Y, Valero L, Lorenzetti ER y Pérez R. 2013. Evaluación de accesiones cubanas de papaya (Carica papaya) ante la mancha anular. Summa Phytopathologica 39(1):24-27. Disponible en línea: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S0258-59362014000300004 [ Links ]
Rodríguez MD, de Sousa GDP, González OJ and dos Reis FA. 2014. Molecular and biological studies of papaya ringspot virus isolates from Brazil and Cuba. American Journal of Agriculture and Forestry 2(5):209-218. DOI: 10.11648/j.ajaf.20140205.11 [ Links ]
Roff MNM. 2007. Disease rating of papaya cultivars to papaya ringspot virus in Malaysia. Acta Horticulturae . 740:277-281. Disponible en línea: http://www.actahort.org/books/740/740_34.htm [ Links ]
Sharma SK and Tripathi S. 2016. Resistance against papaya ringspot virus in Vasconcellea species: present and potential uses. In: Gaur RK, Petrov NM, Patil BL and Stoyanova MI (eds.). Plant Viruses: Evolution and Management. Springer Science Business Media, Singapore. 312p. [ Links ]
Siar SV, Beligan GA, Sajise AJC, Villegas VN and Drew RA. 2011. Papaya ringspot virus resistance in Carica papaya via introgression from Vasconcellea quercifolia. Euphytica 181:159-168. DOI: 10.1007/s10681-011-0388-z [ Links ]
Silva-Rosales L, González-de-León D, Guzmán-González S, and Chauvet M. 2010. Why there is no transgenic papaya in México. Transgenic Plant Journal 4: 45-51. Disponible en línea: http://www.globalsciencebooks.info/Online/GSBOnline/images/2010/TPJ_4(SI1)/TPJ_4(SI1)45-51o.pdf [ Links ]
Silva-Rosales L, Becerra-Leor N, Ruiz-Castro S, Téliz-Ortiz D and Noa-Carranza JC. 2000. Coat protein sequence comparisons of three Mexican isolates of papaya ringspot virus with other geographical isolates reveal a close relationship to American and Australian isolates. Archives of Virology 145:835-843. DOI: 10.1007/s007050050676 [ Links ]
Singh V, Rao GP and Shukla K. 2005. Response of commercially important papaya cultivars to papaya ringspot virus in eastern U.P. conditions. Indian Phytopathology. 58(2):212-216. Disponible en línea: https://www.researchgate.net/profile/Vimla_Singh2/publication/268449279_Response_of_commercially_important_papaya_cultivars_to_papaya_ringspot_virus_in_eastern_UP_conditions/links/546c2aff0cf2397f7831d08a/Response-of-commercially-important-papaya-cultivars-to-papaya-ringspot-virus-in-eastern-UP-conditions.pdf [ Links ]
Sit TL, Abouhaidar MG and Holy S. 1989. Nucleotide sequence of papaya mosaic virus RNA. Journal of General Virology 70:2325-2331. DOI: 10.1099/0022-1317-70-9-2325 [ Links ]
Subramanya SK and Zitter TA. 2014. Plant Virus and Viroid Diseases in The Tropics. Vol. 2: Epidemiology and Management. Springer Netherlands. New Delhi, India. 489p. [ Links ]
Sudha R, Balamohan TN, Soorianathasundaram K, Manivannan N and Rabindran R. 2013. Evaluation of F2 intergeneric population of papaya (Carica papaya L.) for resistance to papaya ringspot virus (PRSV). Scientia Horticulturae 158:68-74. DOI: 10.1016/j.scienta.2013.04.031 [ Links ]
Sudha R, Balamohan TN, Soorianathasundaram K, Rabindran R and Manoranjitham SK. 2015. Studies on resistance to papaya ring spot virus (PRSV) in intergeneric populations of Carica papaya L. and Vasconcellea cauliflora. SABRAO Journal of Breeding and Genetics 47(2): 113-123. Disponible en línea: http://www.sabrao.org/journals/June2015/SABRAO-J-Breed-Genet-47-2-113-123-Sudha.pdf [ Links ]
Taylor D.R. 2001. Virus diseases of Carica papaya in Africa: their distribution, importance, and control. Pp:25-32. In: Hughes JDA and Odu BO (eds.). Plant virology in Sub-Saharan Africa: Proceedings of a Conference Organized by IITA: 4-8 June 2001, International Institute of Tropical Agriculture, Ibadan, Nigeria. Africa. Disponible en línea: http://docplayer.net/35000541-Virus-diseases-of-carica-papaya-in-africa-their-distribution-importance-and-control.html [ Links ]
Tennant PF, Fermin GA and Roye ME. 2007. Viruses infecting papaya (Carica papaya L.): etiology, pathogenesis, and molecular biology. Plant Viruses 1(2):178-188. Disponible en línea: http://www.globalsciencebooks.info/Online/GSBOnline/images/0712/PV_1(2)/PV_1(2)178-188o.pdf [ Links ]
Tripathi S, Suzuki JY, Ferreira SA and Gonsalves D. 2008. Papaya ringspot virus-P: characteristics, pathogenicity, sequence variability and control. Molecular Plant Pathology 9(3):269-280. DOI: 10.1111/j.1364-3703.2008.00467.x [ Links ]
VanBuren R, Zeng F, Chen C, Zhang J and Wai CM. 2015. Origin and domestication of papaya Yh chromosome. Genome Research. 25: 524-533. http://dx.doi.org/10.1101/gr.183905.114 [ Links ]
Veena GL and Dinesh MR. 2013. Utilization of wild species and molecular markers in papaya crop improvement. International Journal of Recent Scientific Research 4(11):1858-1861. DOI: 10.24327/IJRSR [ Links ]
Vegas A, Trujillo G, Sandrea Y y Mata J. 2003. Obtención, regeneración y evaluación de híbridos intergenéricos entre Carica papaya y Vasconcellea cauliflora. Interciencia 28(12):710-714. Disponible en línea: http://www.scielo.org.ve/scielo.php?script=sci_arttext&pid=S0378-18442003001200008 [ Links ]
Villegas VN. 2001. Approaches in developing ringspot virus resistant papaya. Agricultural Sciences Division. Transactions of the National Academy of Science & Technology. 23:63-70. [ Links ]
Yanthan JL, C Vasugi, MR Dinesh, MK Reddy and R Das. 2017. Evaluation of F6 intergeneric population of papaya (Carica papaya L) for resistance to papaya ring spot virus (PRSV) Int. J. Curr. Microbiol. App. Sci 6(5): 289-298. https://doi.org/10.20546/ijcmas.2017.605.033 [ Links ]
Yorobe J M. 2009. Costs and benefits of bioengineered papaya with resistance to papaya ringspot virus in the Philippines. Pp: 23-34. In: GW Norton and DM Hautea (eds.). Projected impacts of agricultural biotechnologies for fruits & vegetables in the Philippines & Indonesia. ISAAA-SEAMEO, Philippines. 184 p. Disponible en línea: https://www.isaaa.org/programs/impact_assessment_of_crop_biotechnology/download/Costs%20and%20Benefits%20of%20PRSV-resistant%20Papaya.pdf [ Links ]
Received: March 16, 2017; Accepted: July 27, 2017











 texto en
texto en 


