Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.35 n.3 Texcoco Sep. 2017
https://doi.org/10.18781/r.mex.fit.1705-1
Scientific articles
Techniques for isolation, identification and molecular characterization of Moko disease-related Ralstonia solanacearum strains
1Unidad de Biotecnología, Centro de Investigación Científica de Yucatán, A.C., Calle 43 No. 130 X 32 y 34, Colonia Chuburná de Hidalgo, C.P. 97205, Mérida, Yucatán, México.
2Centro de Investigación y Asistencia en Tecnología y Diseño del Estado de Jalisco, A.C. Av. Normalistas 800, Colinas de La Normal, C.P. 44270, Guadalajara, Jalisco, México.
Ralstonia solanacearum is one of the most destructive phytopathogens in the agriculture. In banana cultivation, the race 2 causes Moko disease, which is present in some areas in Mexico, and when epidemic outbreaks burst there are losses. Race 2 comprises subgroups (sequevars) which are usually identified by Multiplex PCR reactions. Isolation of R. solanacearum is usually hard, even on semiselective media, because it grows slowly and it is frequently contaminated with Pseudomonas, Klebsiella and Erwinia bacteria, which show similar phenotypes. This work shows a strategy that make easy the isolation, identification and classification of R. solanacearum race 2. The protocol comprises immunodiagnosis of plant suspicious samples, confirmation of diagnosis by PCR, isolation on semiselective media SMSA and B-King, and genotyping of sequevars by single, specific PCR reactions. In the present work 25 strains were isolated from Tabasco, and they genotyped in sequevar 6. Their previous genotyping by Multiplex yielded confuse results, which make evident that Multiplex can result in incorrect conclusions. We discourage to propose genetic divergence and emergency of new strains based on Multiplex results, as done in some recent reports.
Key words: Banana; diagnosis; bacterial isolation; genotyping; sequevar
Ralstonia solanacearum es uno de los fitopatógenos más destructivos en la agricultura; en el cultivo de plátano la raza 2 causa la enfermedad denominada Moko del plátano, la cual en México está restringida a algunas áreas y provoca pérdidas cuando ocurren brotes epidémicos. La raza 2 comprende subgrupos (secuevares), los cuales usualmente son identificados mediante PCR de tipo Multiplex. El aislamiento de R. solanacearum es usualmente difícil, aún sobre medios semiselectivos, debido a su crecimiento lento y a la frecuente contaminación con bacterias Pseudomonas, Klebsiella y Erwinia, que presentan fenotipos similares. En este trabajo se presenta una estrategia que facilita el aislamiento, identificación y clasificación de R. solanacearum raza 2. El protocolo comprende inmunodiagnóstico del material vegetal sospechoso, confirmación mediante PCR, aislamiento sobre medios semiselectivos SMSA y B-King, y genotipificación de los secuevares mediante reacciones de PCR sencillas, específicas. En el presente trabajo se aislaron 25 cepas de muestras obtenidas del estado de Tabasco, las cuales genotipificaron en el secuevar 6. Su previa genotipificación mediante Multiplex fue confusa, lo que evidencia que ese ensayo puede llevar a conclusiones erróneas. Por lo tanto no es válido proponer divergencia genética y emergencia de nuevas cepas con base en su resultado, como se ha hecho en algunos reportes recientes.
Palabras clave: Plátano; diagnóstico; aislamiento bacteriano; genotipificación; secuevar
Ralstonia solanacearum (Smith, 1896) is one of the most destructive bacterial plant pathogens in agriculture. There is a high variability within the species, therefore R. solanacearum is considered a “species complex”, abbreviated as RSSC, for “Ralstonia solanacearum species complex” (Safni et al., 2014; Prior et al., 2016). A species complex is defined as a group of closely-related isolations, the individual members of which can represent more than one species (Fegan and Prior, 2005). R. solanacearum has a wide global distribution and a high number of host plants, which include hundreds of plants from at least 54 botanical families, both dicotyledonous and monocotyledonous (Prior et al., 2016). Many of the plant species affected are strategic crops or important subsistence foods. They include potatoes (Solanum tuberosum), tomato (S. lycopersicum), eggplant (S. melongena), peanut (Arachis hypogea) and banana (Musa spp.) (Cardozo et al., 2010).
Bananas are the most widely produced tropical fruit in the world. In 2013, a worldwide production of approximately 17 million tons was reported (FAO, 2014). India is the main producer, with 26% of the global production. Bananas are a basic staple food and its export is an important source of income and jobs in several developing countries.
Mexico is one of the world’s 15 largest banana producers (FAO, 2017; Workman, 2017), with a production volume of 2.2 million tons and a revenue of MXN$ 6,209,716.85 (SIAP, 2017). It is planted in 17 states of Mexico, with Chiapas and Tabasco as the states with the highest production rate, contributing with 60% of the country´s production (SIAP, 2017).
The bacterial Moko is one of the most important plant health problems in the banana agroindustry in tropical countries, since it is the main bacterial disease in this crop. Currently, the disease is present in several banana-producing countries in the Americas, Asia, Africa, and Oceania (Cardozo et al., 2010).
The pathogen survives or remains latent in infected harvest residues and the rhizosphere of host weeds, the latter being the main inoculant, and infecting healthy plants through the roots and lesions, colonizing the xylem vessels. Symptoms include yellow, flaccid leaves that finally collapse and adhere to the plant; fruits have a gummy texture, with a dark appearance, and tending to degrade. An internal vascular necrosis is present, along with dark dots due to a systematic infection. This disease is common in the genotype ABB (cooking banana), but all types of bananas are affected (Albuquerque et al., 2014). In Mexico, this disease appears in some areas and is subjected to official control; the campaign against banana Moko operates in the states of Chiapas, Tabasco, and Nayarit in order to reduse the spreading of the disease into areas without it (SENASICA, 2016).
Reports on R. solanacearum, the cause of Moko, claim it was introduced into Mexico in 1960 in the banana-producing area of Tapachula, Chiapas. Eight years later, the disease spread into another 12 municipal areas of the same state. In 1991, the first reports were presented in Teapa, Tabasco, and in 2009, the disease spread to the areas of Cunduacán, Huimanguillo, Centro, Cárdenas, and Jalapa. The banana Moko is present in the banana-producing municipal areas of Pichucalco, Huehuetán, Mazatán, Suchiate, and Tapachula in the state of Chiapas; in the municipal areas of Centro, Cunduacán, Huimanguillo, Jalapa, Teapa, and Cárdenas in the state of Tabasco, and in Santiago Ixcuintla in the state of Nayarit. Its incidence has increased in the state of Tabasco in recent years (SENASICA, 2015). It is currently under official control, with the plant health campaign against the Moko disease of banana, which is operant in the states of Chiapas, Tabasco, and Nayarit, with the aim of reducing the risk of spreading to areas with no incidence, reducing infestation levels in the areas under phytosanitary control (SENASICA 2017).
R. solanacearum is subdivided into five biovars, based on its carbohydrate metabolism, and five races, based on the range of hosts it infects (Denny and Hayward, 2001; Fegan and Prior, 2005; Champoiseau et al., 2009). Races can be host-specific, such as races 4 and 5, which infect ginger and blackberry, respectively, or they can present a wide range, such as race 1 (tomato, potato, eggplant, and others), race 3 (potato, tomato, and geranium), and race 2 (heliconia and bananas) (Denny and Hayward, 2001; Champoiseau et al., 2009).
Since R. solanacearum makes up a heterogenous group of strains with a high genetic and phenotypic diversity, its classification has been under constant reconsideration for 50 years (Hayward, 1964; Fegan and Prior, 2005; Safni et al., 2014; Prior et al., 2016). Classification into biovars does not reflect the genetic background of the strains, so a genetic classification scheme is currently used, hierachically organized into the categories of phylotypes and sequevars (Fegan and Prior, 2005; Prior et al., 2016). Phylotypes correlate the strains of R. solanacearum with its geographic origins: phylotype I comprises mostly Asian strains; phylotype II, strains from the Americas; phylotype III, from Africa and the Indian Ocean; and phylotype IV, from Indonesia, Japan and Australia (Fegan and Prior, 2005). Phylotypes are subdivided into sequevars, which are based on the nucleotidic sequence of the gene Egl, which codifies for an endoglucanase (Fegan and Prior, 2005). So far, 54 sequevars have been identified in the complex R. solanacearum (Li et al., 2016); the sequevars that cause the Moko disease in bananas are 3, 4, 6, 24, 25, 41, and 53. In the Americas, the Moko disease is present from Brazil to the United States (Sanchez-Perez et al., 2008; Norman et al., 2009; Hong et al., 2012; Albuquerque et al., 2014).
For genetic studies, pathogenesis, or other characterizations, R. solanacearum is usually isolated in the semiselective media SMSA (“selective media from South Africa”), (Álvarez-Restrepo et al., 2008; Cardozo et al., 2010; Elphinstone et al., 1996). However, even in this medium, isolating this bacteria is difficult, due to the growth of other microorganisms (Champoiseau et al., 2009; Kalpage y De Costa, 2015), particularly bacteria of the genera Klebsiella, Erwinia, and Pseudomonas. These three gram-negative bacteria, like other species of Ralstonia, such as Ralstonia mannitolilytica, are majority endophytic in banana plants (Thomas et al., 2008; Ganen et al., 2009; Souza et al., 2013). These bacteria are common contaminants in R. solanacearum plantations, since they present a faster growth and similar microscopic and colonial morphologies (French et al., 1995; Nasim, 2011).
The isolation of pure R. solanacearum strains can take a long time (French et al., 1995; Thera, 2007; Döölotkeldieva and Bobuşeva, 2014; Döölotkeldieva and Bobusheva, 2016). This report describes a strategy that makes the isolation and genetic characterization of R. solanacearum easier, from banana diseased with Moko.
MATERIALS AND METHODS
Sampling plant tissues
Samples were taken in Tabasco (October 20-24, 2014) with the technical support of the personnel from the Tabasco Local Plant Health Committee (Comité Local de Sanidad Vegetal de Tabasco - CESVETAB) for the location, visual diagnosis and collection of material, according to instructions by SENASICA. The samples were deposited in Ziploc bags and labelled with the information of the tissue collected, date and sampling site. The samples were places in coolers with cooling gels and sent to the banana molecular biotechnology lab in the Scientific Research Center (Centro de Investigación Científica) in Yucatan, where they were processed.
Detection of R. solanacearum by immunology tests
To detect R. solanacearum, the ELISA (“Enzyme-Linked Immunosorbent Assay”) (Agdia®, N.C. SRP 33900) was carried out in a 96-well tray, following the instructions of the manufacturer. Positive controls and samples of plants infected with R. solanacearum produce a blue color, whereas negative controls and samples of healthy plants produce a colorless result. The tray was visually examined to avoid false positives by possible contaminations or impurities on the tray and the spectrophotometric absorbance was measured at 655 nm (Bio-Rad Modelo 680XR). Optic densities equal to or lower than 0.09 was considered negative. To scrutinize the samples in the lab, ImmunoStrips (Agdia®, N.C. STX33900) were also used, following the instructions of the manufacturer.
DNA extraction
To enrich the R. solanacearum population in the SMSA medium, a fragment (1 cm2) of the infected plant tissue was deposited in the liquid medium and incubated for 24 h, shaking constantly at 180 rpm at 28 °C (Álvarez-Restrepo et al., 2008). One aliquot of this culture (1.5 mL) was centrifuged at 5,200 X g for 5 min. The bacterial pellet was resuspended with 1 mL of the buffer CTAB (2% cetyl trimethyl ammonium bromide, 1% polyvinylpyrrolidone, 100 mM Tris-HCl, 1.4 M NaCl, 20 mM EDTA), stirring with a vortex for 30s. Later it was frozen at -20 °C for 10 min and incubated at 65 °C in a water bath for 20 min. It was tempered for 1 min at room temperature and centrifuged at 20,800 X g. The supernatant was recovered in a 1.5 mL tube for centrifuging, 500 µL of phenol:chlorophorm:isoamylic (25:24:1) were added, and stirred for 10 min at room temperature (CNRF, 2012). Later, it was centrifuged for 5 min at 20,800 X g and the acqueous phase was collected. Next, the standard DNA recovery procedure was carried out using cold absolute ethanol, washings with 70% ethanol, air-drying of the pellet and resuspension in ultrapure distilled water (Sambrook and Russell, 2001). The DNA purity was determined by spectrophotometry at 260nm/280nm and the integrity was determined by electrophoresis in agarose gel.
Detection of R. solanacearum by PCR
Detection was carried out by the amplification of genomic regions with the primers of 759: 5’- GTC GCC GTC AAC TCA CTT TCC - 3’ and 760: 5’- GTC GCC GTC AGC AAT GCG GAA TCG- 3 (Universal diagnosis for R. solanacearum, 280 pb), (Opina et al., 1997); pehA#3-F 5’-CAG CAG AAC CCG CGC CTG ATC CAG- 3’ and pehA#6-R 5’- ATC GGA CTT GAT GCG CAG GCC GTT- 3’ (gen pga, 532pb), (Gillings et al., 1993); ISRso19-F 5’- TGG GAG AGG ATG GCG GCT TT - 3’ and ISRso19-R 5’ - TGA CCC GCC TTT CGG TGT TT - 3’ (primers specific to Race 2; 1884pb), (Lee and Chin, 2003). The PCR reaction mixture was prepared with 20ng of DNA, 0.1 µM of each primer, 1 U of recombinant DNA Taq polymerase (Invitrogen®), IX of the Buffer of the Taq enzyme, 200 µM of each dNTP and 1.5 mM MgCl2, in 25µL of final volume. The program in the thermocycler (Bio-Rad model T100™) for the universal diagnosis of R. solanacearum (primers 759/760) had a denaturalization cycle of 2 min at 96 °C, followed by 35 cycles of 94 °C for 45 s, alignment step at 64 °C for 20 s, and a step of 72 °C for 1 min, and finally, an extension cycle of 72 °C for 5 min. The amplification program for the other genes was identical, except for the step of alignment was 70 °C for 45 s for the gene pga, and for Race 2, alignment was 55°C for 30 s, and the extension of 72 °C for 1.5 min in each cycle. The products (10 µL of each PCR reaction) were analyzed by electrophoresis in 1.5% agarose gel (p/v) using the buffer TAE (Tris-acetate-EDTA) 1X supplemented with 3 µL of ethidium bromide (10mg/mL). The strips were visualized over a transilluminator using UV light (Gel Doc™ XR, Bio-Rad).
Isolation of R. solanacearum
Tissues (pseudostem and fruit) which were positive at immunology tests ELISA and ImmunoStrips were used for bacterial isolation. Samples were cut into small pieces with a sterilized blade, and later disinfested by washing twice with sterilized water, sodium hypochlorite at 10% (from a commercial solution at 6%) for 1 min, sterile water, ethanol for 30 s, sterile water, and it was later placed in TE buffer and left to incubate for an hour, stirring softly. A bacterial loop was placed on a semi-selective SMSA medium (formula for every liter: 1g of casamino acids, 10g of peptone, 5g of glucose, 5mg crystal violet, 100mg polymyxin β-sulfate, 25mg bacitracine, 5mg chloranphenicol, 0.5mg penicillin, 17g bacteriological agar, 50mg TZC (2,3,5-triphenyl tetrazolium chloride) and incubated at 28 °C for 72 h (Kelman, 1954; Cardozo et al., 2010). The bacterial colonies were streak by crossing on a solid B-King medium and incubated at 28 °C for 48 h. The B-King medium helps to distinguish Pseudomonas spp. bacteria, since they produce fluorescence when exposed to a UV wavelength of 360 nm, whereas R. solanacearum does not fluoresce (King et al., 1954; Dulla and Lindow, 2009; Lamichhane and Varvaro, 2013).
Caracterization of the sequevar
For the characterization of the sequevars, we used the primers of the Mus serie from Fegan and Prior (2005): 5´-CGGGTCGCTGAGACGAATATC-3´ and 5´-GCCTTGTCCAGAATCCGAATG-3´ (Sequevar 4 , 351 pb); 5´-GCAGTAAAGAAACCCGGTGTT-3´ and 5´-TCTGGCGAAAGACGGGATGG-3´ (Sequevar 3, 400 pb); 5´-GCTGGCATTGCTCCCGCTCAC-3´ and 5´-TCGCTTCCGCCAAGACGC-3´ (Sequevar 4 SFR, 167 pb); 5´-CGTTCTCCTTGTCAGCGATGG-3´ and 5´-CCCGTGTGACCCCGATAGC-3´ (Sequevar 6, 221 pb). PCR was carried out in Multiplex using 20 ng of DNA, 2 U of recombinant DNA Taq polymerase (Invitrogen®), 1X of the Buffer of the Taq enzyme, 200 µM of each dNTP, 1.5 mM MgCl2 and 6 pmoles of each primer, in a final volume of 25µL. The PCR conditions were similar to those described previously, using an alignment temperature of 59 °C. Single PCRs for each sequevar were carried out in the same conditions, but using only one pair of primers in each case. In each case, 15 µL of the product of the reaction was analyzed by in gel electrophoresis.
RESULTS AND DISCUSSION
The diagnosis for bacterial Moko disease was carried out on suspicious banana plant pseudostems and fruits. The diagnosis of the plant material collected was carried out with 2 serological tests and 2 PCR tests. A total of 40 samples were analyzed, out of which 35 were positive in the diagnosis with ELISA and 28 with the diagnosis with ImmunoStrips. All 40 were analyzed with PCR with the two pairs of primers, and both PCR reactions amplified 28 samples. The diagnosis with the ImmunoStrips provided a congruence of 100% with the PCR diagnoses, both in the number of positives and in the samples identified, whereas the ELISA test presented 20% of error (7 false positives out of the 35). Neither of the two immunological methods presented false negatives (Table 1). Figures 1A and 1B show typical results obtained in the ELISA diagnoses and with ImmunoStrips. Some ImmunoStrips were darker than others, depending on the phenolization of the extract, yet the color did not interfere in the reaction of the control line and that of the negative or positive result.
Table 1 Diagnosis of the bacterial Moko disease in banana plan ts with suspicious symptomatology.
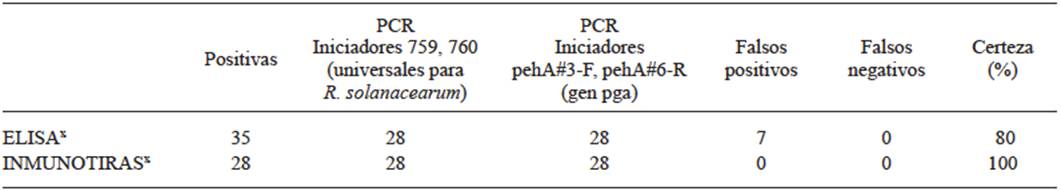
x A total of 40 samples were analyzed: 14 pseudostem and 26 fruit samples.
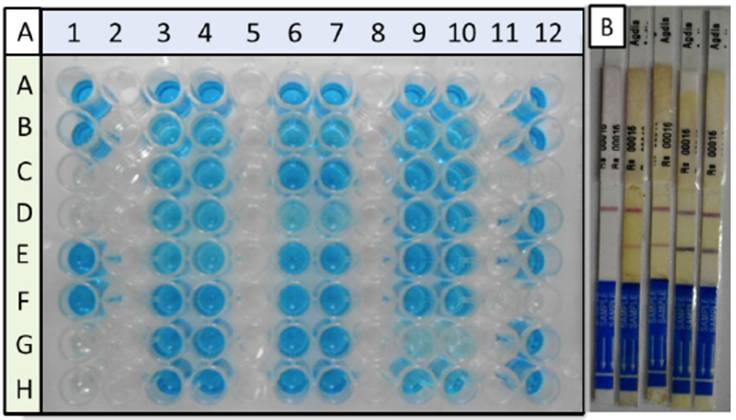
Figure 1 Serological diagnosis of bacterial Moko disease in banana pseudostem and fruit samples. Panel A, diagnosis by ELISA. Controls and samples were analyzed by duplicate. Column 1, negative (Rows C, D, G, and H) and positive controls (Rows A, B, E, and F). Samples were placed in columns 3, 4, 6, 7, 9, 10, and 12. Panel B, diagnosis by immunostrips; the result of a negative sample is included (only one line for the control); positive samples, two lines. Both diagnoses show differences in the intensity in color of the signal in the positive samples, which is related to the differences in the amount of inoculant, according to the manufacturer.
The ELISA technique has been widely used in the protocols for the identification of R. solanacearum amongst colonies with similar phenotypes (Álvarez-Restrepo et al., 2008; Döölotkeldieva and Bobusheva, 2016). The diagnosis is based on the use of monoclonal antibodies against exopolysaccharides (EPS) produced by the bacteria. Naranjo-Feliciano and Martínez-Zubiaur (2013) diagnosed diseased plant tissues by comparing the ELISA trials, agglutination with latex particles, and the detection of R. solanacearum by PCR. The agglutination with latex particles was the quickest and simplest technique of the 3, but it was less specific, since it reacted with other microorganisms, and it is less sensitive by one order of magnitude (límite 106 ufc/mL) than the other serological test. According to the manufacturer, the R. solanacearum detection limit in the ELISA technique is of 105 ufc/mL. These authors observed differences in reliability of the detection of the pathogenic bacteria by the 3 methods, when they compared the results of the diagnosis with the development of the plant’s disease, afterwards. The reliability of the diagnosis using PCR was up to 20% higher than with the ELISA test. In this work, the PCR test confirmed 80% of the positive cases diagnosed using ELISA, indicating 20% of false positives in this serological tests, which coincides with the report by Naranjo-Feliciano and Martínez-Zubiaur (2013). Other authors have discussed the limitations of the ELISA test and the advantages of molecular markers, which can detect up to 102 ufc/mL (Álvarez-Restrepo et al., 2008). Eriksson and Aspan (2007) compared PCR and ELISA in the diagnosis of bacteria of the genus Salmonella in pig fecal samples, and they found discrepancies between the results of both techniques due to the low specificity of the antibodies used, which shows that the problem of the immunodiagnosis by ELISA is not exclusive of the R. solanacearum diagnosis.
The R. solanacearum diagnosis in the banana samples with ImmunoStrips was 100% congruent with the later detection by PCR. ImmunoStrips are a pocket diagnosis, also sold by Agdia®, Inc. and offers the same limit of detection than the ELISA test (105 ufc/mL). These ImmunoStrips have been used in other investigations as a method to confirm the identities of isolated R. solanacearum strains (Sánchez-Pérez et al., 2008; Hong et al., 2012). Thera (2007) used ImmunoStrips to diagnose R. solanacearum in tomato, potato, tobacco, and green chili pepper tissues. The ImmunoStrips had one false negative in a frozen sample of green chilies and confirmed 24 of the 25 infected samples (96%), supporting the fact that this test has a high degree of certainty.
Korus (2011) diagnosed Clavibacter michiganensis comparing ELISA and ImmunoStrips and obtained similar results of an efficiency of 100% using ImmunoStrips, in comparison with variable detection efficiencies (33 to 100%) in ELISA tests for different pathogen subspecies. Both tests can use the same antibodies, as in the case of Agdia® products to detect R. solanacearum. Detection with ImmunoStrips is simple, i.e. the sample is macerated and applied on the ImmunoStrip, whereas ELISA tests include multiple steps such as buffer preparations, prolonged incubation times, and multiple washing steps, which increase the possibilities of error. A dilution effect could also occur in the sample in the ELISA test, affecting the detection in some samples (Ruiz-García et al., 2009).
The robustness of the R. solanacearum diagnosis with ImmunoStrips in banana tissues in this and other investigations (Thera, 2007), as well as its speed, simplicity and portability, make its use advisable to monitor R. solanacearum on the field, and for the gathering of samples for the isolation and study of the bacteria. The confirmation of the diagnosis must continue to be with molecular markers, since the limits of serological detection do not allow for the detection of cases of early infection (Thera, 2007), nor do they distinguish between the different races of R. solanacearum (Rajeshwari et al., 1998).
Isolation of R. solanacearum
With the sequential use of 2 culture media, SMSA and B-King, it was possible to isolate 25 R. solanacearum strains from the 28 positive samples. The three samples from which R. solanacearum could not be recovered had the palest diagnosis lines in the ImmunoStrips, indicating that these samples contained a low bacteria count.
The SMSA medium is frequently used to isolate R. solanacearum, since it avoids the growth of many other microorganisms (Kelman, 1954; Álvarez-Restrepo et al., 2008; Cardozo et al., 2010). However, even so, R. solanacearum is difficult to isolate since it has slow growth rates in vitro, while other bacteria, e.g. Pseudomonas, grow quickly and contaminate the isolations (Ramesh et al., 2009; Champoiseau et al., 2009; Nasim, 2011; Kalpage and De Costa, 2015). The combination of the media SMSA and B-King helps to distinguish R. solanacearum from Pseudomonas spp. Both of these bacteria grow on the SMSA medium as mucoid cultures, with irregular, white edges, and pink to red centers. On the B-King medium, Pseudomonas spp. fluoresces, whereas R. solanacearum do not (Figure 2), helping to put them apart. Ramesh et al., (2009) point out that genus Pseudomonas is opportunist during the colonization of the host by R. solanacearum. Along with Pseudomonas, the bacteria Klebsiella, Erwinia, and other species of the genus Ralstonia, such as Ralstonia mannitolilytica, are mostly endophytic of the banana (Thomas et al., 2008; Ganen et al., 2009; Souza et al., 2013). The bacteria recovered from the 3 samples that produced faint lines on the ImmunoStrips presented fluorescence on the B-King medium and did not amplify in any of the PCR diagnosis reactions for R. solanacearum. Some of these bacteria were identified by sequencing the 16S, and belonged to Pseudomonas fulva, Pseudomonas aeruginosa, and Pseudomonas citronellolis, consistent with reports by the bibliography.
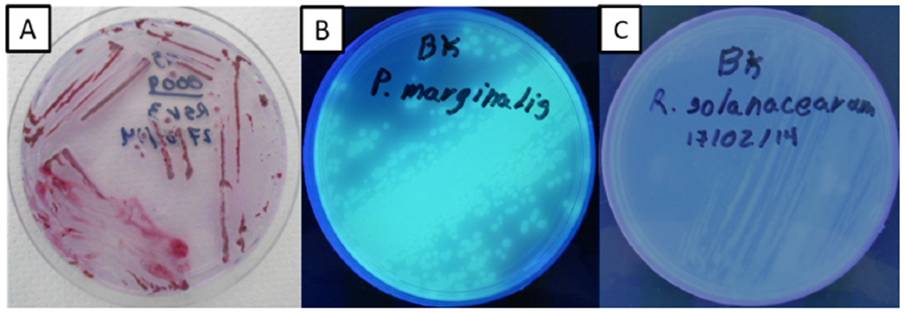
Figure 2 Preselection of bacteria with similar phenotypes to R. solanacearum. Isolation on SMSA medium (panel A). Strains with mucoid characteristics, white, irregular edges and red centers were transferred on a B-King medium. Bacteria of the genus Pseudomonas fluoresce on this medium (Panel B). Non-fluorescent bacteria were selected (panel C) to continue the isolation of R. solanacearum.
Confirmation of the identity of R. solanacearum
The identity of the R. solanacearum strains was confirmed by sequencing the amplified fragment with primers pehA#3-F, pehA#6-R (fragment of the gene pga). The 25 strains that amplified the 532pb diagnostic band were confirmed to be R. solanacearum using the BlastN analysis with the sequences obtained, and compared with the GenBank database (hit 99-100% identity with R. solanacearum). Based on the positive PCR results when using the primers ISRso19, the 25 isolated strains corresponded to Race 2.
The identification of the sequevars was first determined by PCR Multiplex, using the primers of the series Mus (Fegan and Prior, 2005). According to these authors, each sample must give only one diagnostic DNA band: ~167 pb (sequevar 4 ecotype SFR), ~350 pb (sequevar 4), ~220 pb (sequevar 6), ~400 pb (sequevar 3). These DNA bands were observed among the products of the Multiplex (Figure 3A), although they were not unique DNA bands per sample. According to the Multiplex (Figure 3A), the Race 2 R. solanacearum strains genotype in sequevar 3 (lanes 11 and 13), sequevar 4 (lanes 3 and 5), sequevar 4 SFR (lane 1), and sequevar 6 (lanes 2, 4, 6, 8). Other strains produced profiles that could not be assigned to any of the sequevars (e.g. lanes 7, 10). Due to the inconsistency in results of the PCR in Multiplex, each strain was later genotyped using independent PCR for each sequevar. No Tabasco strains amplified with the pairs of primers for sequevars 3, 4 and 4 SFR (data not shown). The 25 strains amplified the diagnostic DNA band of ~220 with the pair of primers SI28, that corresponds to sequevar 6 (Figure 3B). Sequevar 6 originated in the Carribbean islands (Trinidad) and entered the continent mainland through Venezuela; in 1950, it was accidentally introduced into Costa Rica in infected banana corms. In 1960, a commercial freight ship introduced it accidentally into Honduras, and from there it spread rapidly northward to Mexico and southward to Panama (Sequeira, 1998). The first report in Mexico was in 1960 in Chiapas. Currently, sequevar 6 has been reported in North America in Hawaii (Fegan and Prior, 2005) and Florida (Hong et al., 2012); in Central America, in Guatemala (Sánchez-Pérez et al., 2008), and in South America, in Brazil (Alburquerque et al., 2014). Central and South America are considered the center of origin of strains related to the Moko disease of banana (Sequerira, 1998; Fegan and Prior, 2005, 2006). Other countries in the Americas have, along with sequevar 6, some of the sequevars 3, 4, 24, 25, 41, and 53 (Raymundo et al., 1998; Prior and Fegan. 2005; Alburquerque et al., 2014). Studies in Mexico must be broadened to determine if there are other sequevars related to the bacterium causing Moko disease in banana.
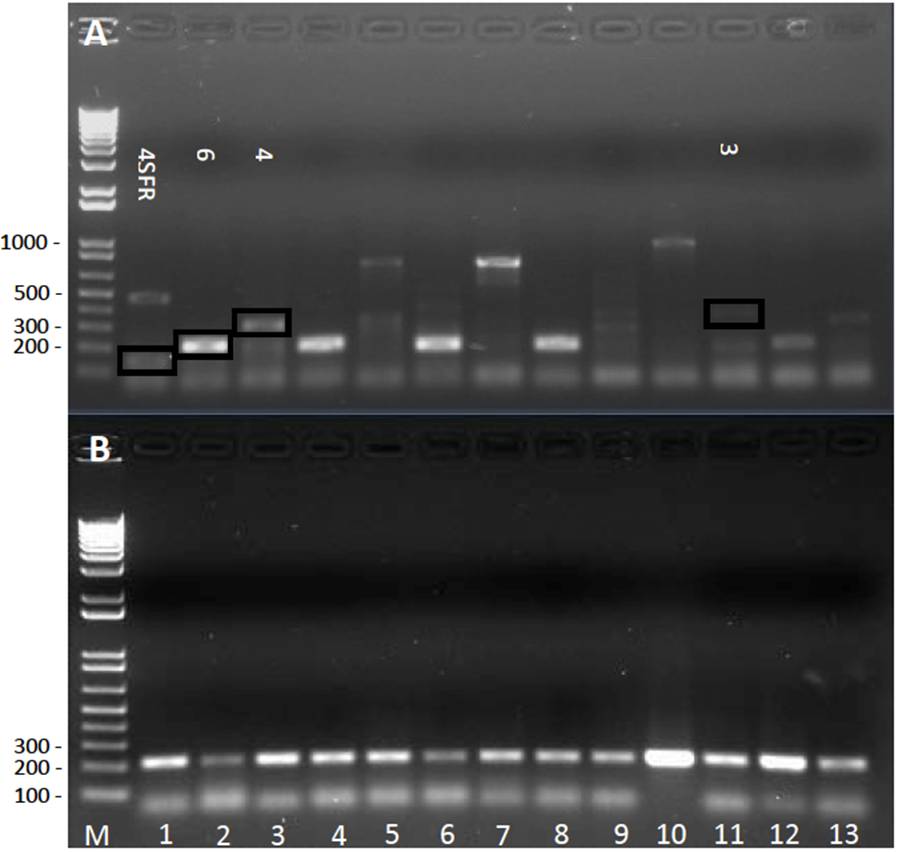
Figure 3 Characterization of R. solanacearum bacteria isolated from banana plants; tipification of the sequevars. Panel A, PCR in Multiplex (primers of the series Mus and SI28 together). Lane 1 shows a band of ~167 pb that corresponds to expected for sequevar 4 SFR, and another of ~450 pb, not described for any sequevar; lane 2 shows a band of approximately ~220 pb that corresponds to descriptions for sequevar 6; lane 3 showed a band of ~350 pb, which corresponds to sequevar 4. The following lanes showed unexpected bands, such as ~850 pb (lanes 5 and 7) and ~1000pb (lane 10). Panel B, PCR with specific primers for sequevar 6 (primers SI28, diagnosis band of ~220 pb). In both panels the lanes correspond to: M) Marker 1 Kb Plus DNA Ladder (Invitrogen® Carlsbad, CA, USA); lanes 1-13, strains of R. solanacearum, 1) MT01, 2) MT02, 3) MT03, 4) MT04, 5) MT05, 6) MT06, 7) MT07, 8) MT08, 9) MT09, 10) MT10, 11) MT11, 12) MT12, 13) MT13.
The taxonomic classification of the strains of the R. solanacearum species complex (RSSC) has been under constant revision and correction for over 50 years, and more frequently in the last 10 years (Peeters et al., 2013; Prior et al., 2016). As the world collections increase, and along with them, the known global genetic diversity of the RSSC, discrepancies have been found in the classification results of some strains with different diagnosis tools (Hong et al., 2012), and even in the classification of populations, such as in the cases of the isolated R. solanacaerun of Solanaceae and Cucurbits in the French Guyana, and tomatoes in Guadalupe (Álvarez-Restrepo et al., 2008; Deberdt et al., 2014). Although the reasons for the inconsistencies are not entirely clear, some of the differences have turned into diagnosis tools. For example, many strains of sequevar 4 produce a 351 pb band with primers Mus06, but they additionally amplify a band of 167pb (Álvarez-Restrepo et al., 2008), while some strains of sequevar 4 isolated in Florida in 2008 do not produce the diagnosis band of 351pb, but only the unspecific 167pb band, therefore the authors propose that the Florida strains that only amplify the 167pb band are emerging strains (Hong et al., 2012).
The Multiplex to distinguish between sequevars 3, 4, and 6 within Race 2 (i.e. strains causing banana Moko) was generated by Fegan and Prior (2005) and it has been used to classify strains in Martinique (Wicker et al., 2007, 2009), Colombia (Álvarez-Restrepo et al., 2008), United States (Hong et al., 2012), French Guyana (Deberdt et al., 2014), Brazil (Albuquerque et al., 2014), and others. In the reports that show electrophoresic analyses of the PCR products, we can notice unspecific bands in some lanes, alongside the diagnosis band (Álvarez-Restrepo et al., 2008; Hong et al., 2012; Albuquerque et al., 2014), as observed in this investigation about Mexico. Even the analysis of one same sequevar shows discrepancies between the different research groups. Sequevar 4 amplifies 2 bands in the Moko Multiplex, although the band sizes are different between the report by Álvarez-Restrepo et al., (2008) and the one by Albuquerque et al., (2014), despite both investigations having used agarose gels with the same concentrations (1.5% p/v).
In this investigation the diagnosis by Multiplex was ambiguous. In the case of the strains that amplified the 221pb band (sequevar 6), it was a single band (Figure 3A, lanes 2, 4, 6, 8, 12), contrary to results by Albuquerque et al., (2014) who obtained 2 bands for the strain IBSBF2661 (sequevar 6), but similar to results obtained by Hong et al., (2012), who also obtained one band for strain 527 (sequevar 6) in the Moko Multiplex. In some Mexican strains, the diagnostic band expected for sequevars 3 (400pb, lane 11) and 4 (351pb, 167pb, lanes 1 and 3) were amplified, although other bands observed in those same lanes do not coincide with any of the reports. PCR bands were also observed that have not been reported previously in the Multiplex results for Moko strains (Figure 3A, lanes 5, 7, 10), or in other words, they are new bands. The new ~850 pb band amplifies in different strains (Figure 3A, lanes 5 and 7), suggesting they have a genetic origin, and it reflects the diversity in the populations of R. solanacearum. These results are difficult to interpret in further detail because the diagnosis primers amplify marker bands, although the function in the microorganism of those genomic regions is unknown (Deberdt et al., 2014).
The presence of new and reproducible PCR products in different strains (e.g. ~850 pb band in strains MT05 and MT07) could be interpreted as an emergence of new strains of R. solanacearum in Tabasco, as proposed for the Florida strains that presented atypical results in this Multiplex diagnosis (Hong et al., 2012). Due to the ambiguity of results, the individual diagnosis of each sequevar was carried out. No amplification of any strain was obtained in the diagnosis of sequevars 3 or 4 (data not shown). All the strains isolated in Tabasco in this investigation belong to sequevar 6 (Figure 3B). The recommendation by the present report is to avoid basing only in Moko Multiplex results to propose the emergence of new R. solanacearum strains. We suggest amplified products to be sequenced.
Figure 4B presents a summary of the protocol proposed from the present investigation, in order to simplify the isolation of R. solanacearum Race 2 strains. It is compared with the procedure followed by other investigations (Figure 4A). Procedure 4A is more expensive, since the diagnosis by ELISA or ImmunoStrips must be carried out on each isolated bacterial strain. In the present research, that strategy had little success. When the samples were not diagnosed before isolation, a large part of the bacteria isolated on the SMSA were not R. solanacearum. The sequential use of B-King medium after the isolation on SMSA is a quick and inexpensive option that allows the phenotypic preselection of R. solanacearum. According to the bibliography, the most reliable R. solanacearum identity diagnoses are PCR and sequencing, whether of ITS or specific genes (Fegan and Prior, 2005). After confirming the identity of R. solanacearum, the collection of isolated bacteria can be characterized genetically, phenotypically, phylogenetically, or be used for other studies. We recommend performing individual PCR with the specific primers of each Moko sequevar instead of the Moko Multiplex whenever results are unclear.
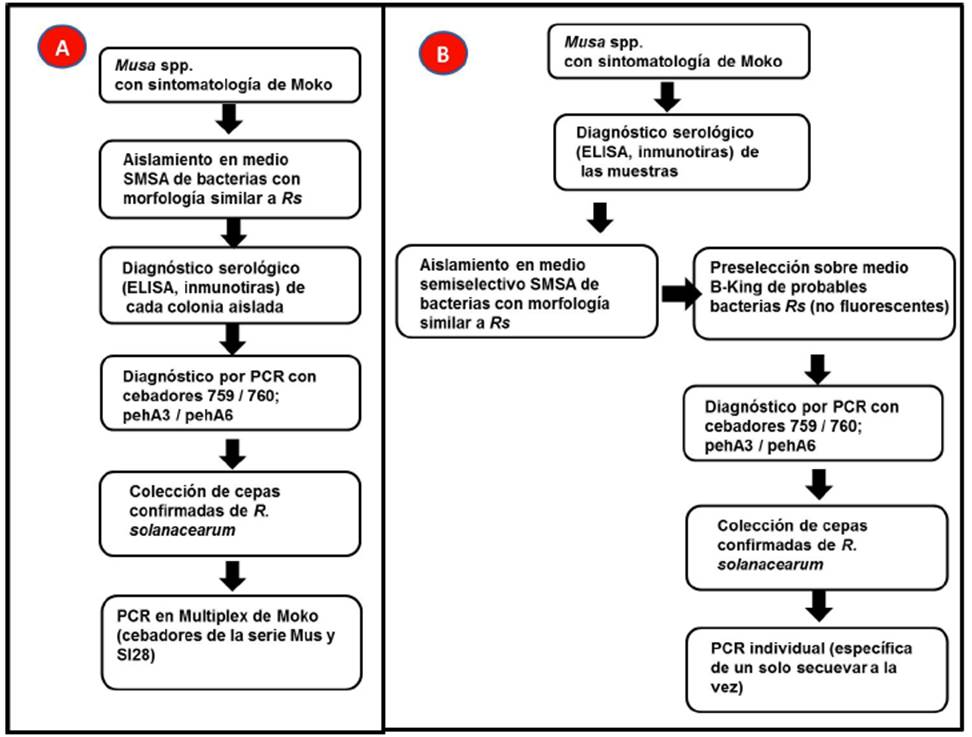
Figure 4 Schematic comparison of the protocol generally used for the isolation of R. solanacearum related to the Moko disease (Panel A), and the protocol recommended in the present study (panel B). Panel A is based on Álvarez- Restrepo et al. (2008); Dööolotkeldieva and Bobusheva (2016); Hong et al. (2012); Sánchez-Pérez et al. (2008).
Moko is a quarantine-regulated disease in Mexico and a phytosanitary campaign is established. Identified outbreaks are therefore under continuous surveillance and eradication measures. It is important to emphasize that due to regulations and for food security, it is crucial to prevent the dispersal of the pathogen by collection of the material by inexperienced personnel. For this reason, all studies of this pathogen in Mexico should be carried out with the permission of, and in collaboration with the General Plant Health Board (Dirección General de Sanidad Vegetal - DGSV), the National Food Health, Safety and Quality Service (Servicio Nacional de Sanidad, Inocuidad y Calidad Agroalimentaria - SENASICA) and the State Plant Health Committees (Comités Estatales de Sanidad Vegetal).
CONCLUSIONS
The systematic use of ImmunoStrips is recommended to strengthen the diagnosis of R. solanacearum in the field.
The isolation and identification of R. solanacearum is difficult, as other authors have mentioned. The modification of the procedure published in the bibliography for the isolation of R. solanacearum facilitated the bacterial isolation from diseases banana plants: the serological diagnosis of the suspicious samples, the systematic inclusion of the B-King medium to discard bacteria of the genus Pseudomonas, and the confirmative diagnosis by PCR with primers pehA#3-F-pehA#6-R, and ISRso19F-ISRso19R.
The detection of atypical results in the Multiplex PCR for Moko disese of banana must be interpreted with care. The results must be confirmed using individual (specific) PCRs for each sequevar.
REFERENCES
Albuquerque GMR, Santos LA, Felix KC, Rollemberg CL, Silva AM, Souza EB, Cellier G, Prior P and Mariano RL. 2014. Moko disease-causing strains of Ralstonia solanacearum from Brazil extend known diversity in paraphyletic phylotype II. Phytopathology 104:1175-1182. http://dx.doi.org/10.1094/PHYTO-12-13-0334-R [ Links ]
Álvarez-Restrepo J, Rodríguez-Gaviria P y Marín-Montoya M. 2008. Detección molecular de Ralstonia solanacearum en agrosistemas bananeros de Colombia. Tropical Plant Pathology 33:197-203. http://dx.doi.org/10.1590/S1982-56762008000300004 [ Links ]
Cardozo C, Rodríguez P, Cotes JM y Marín M. 2010. Variabilidad genética de la bacteria Ralstonia solanacearum (Burkholderiales: Burholderiaceae) en la zona bananera de Urabá (Colombia). Genetic variability of the bacterium Ralstonia solanacearum (Burkholderiales: Burholderiaceae) in the banana-growing region of Uraba (Colombia). Revista de Biología Tropical 58:31-44. http://dx.doi.org/10.15517/rbt.v58i1.5192 [ Links ]
Champoiseau PG, Jones JB, and Allen C. 2009. Ralstonia solanacearum race 3 biovar 2 causes tropical losses and temperate anxieties. Disponible en línea: Plant Health Progress http://dx.doi.org/10.1094/PHP-2009-0313-01-RV [ Links ]
CNRF, 2012. Protocolo de diagnóstico de Ralstonia solanacearum raza 2 (filotipo II) causante del Moko del plátano. SAGARPA-SENASICA. Estandarizado en proceso de revisión. Dirección General de Sanidad Vegetal - Centro Nacional de Referencia Fitosanitaria. México. 30p. [ Links ]
Denny T and Hayward AC. 2001. Laboratory Guide for Identification of Plant Pathogenic Bacteria. American Phytopathological Society Press. St. Paul, Minn., USA. 174p. [ Links ]
Deberdt P, Guyot J, Coranson-Beaudu R, Launay J, Noreskal M, Riviere P, Vigne F, Laplace D, Lebreton L and Wicker E. 2014. Diversity of Ralstonia solanacearum in French Guiana expands knowledge of the “emerging ecotype”. Phytopathology 104:586-596. http://dx.doi.org/10.1094/PHYTO-09-13-0264-R [ Links ]
Döölotkeldieva T and Bobuşeva S. 2014. Identification and prevalence of Ralstonia solanacearum from potato fields of Kyrgyzstan. Journal of Agriculture and Life Science 4:1-9. Disponible en línea: http://journals.manas.edu.kg/mjavl/index.php/mjavl/article/view/11/11 [ Links ]
Döölotkeldieva T and Bobusheva S. 2016. Identification of Ralstonia solanacearum in Kyrgyzstan’s potato fields and the possibility of using biocontrol agents against this pathogen. International Journal of Environmental y Agriculture Research 2(5):146-155. Disponible en línea: http://ijoear.com/Paper-May-2016/IJOEAR-MAY-2016-42.pdf [ Links ]
Dulla GFJ and Lindow SE. 2009. Acyl-homoserine lactone-mediated cross talk among epiphytic bacteria modulates behavior of Pseudomonas syringae on leaves. The International Society for Microbial Ecology Journal 3:825-834. http://dx.doi.org/10.1038/ismej.2009.30 [ Links ]
Eriksson E and Aspan A. 2007. Comparison of culture, ELISA and PCR techniques for salmonella detection in faecal samples for cattle, pig and poultry. BMC Veterinary Research 3:21 http://dx.doi.org/10.1186/1746-6148-3-21 [ Links ]
Elphinstone JG, Hennessy J, Wilson JK and Stead DE. 1996. Sensitivity of different methods for the detection of Pseudomonas solanacearum in potato tuber extracts. EPPO/OEPP Bulletin 26:663-678. Disponible en línea: https://www.researchgate.net/publication/229943089_Sensitivity_of_different_methods_for_the_detection_of_Ralstonia_solanacearum_in_potato_tuber_extracts [ Links ]
FAO, 2014. Banana market review 2013-2014. Banana Information Note 2014 8p. Disponible en línea: http://www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Bananas/Documents/Banana_Information_Note_2014-_rev.pdf [ Links ]
FAO, 2017. Banana statistical compendium 2015-2016. 26p. Disponible en línea: http://www.fao.org/3/a-i7409e.pdf [ Links ]
Fegan M and Prior P. 2005. How complex is the “Ralstonia solanacearum species complex”. Pp 1-15. In: Allen C, Prior P, Hayward AC (eds). Bacterial wilt disease and the Ralstonia solanacearum species complex. Minnesota, American Phytopathological Society Press. St. Paul, Minn., USA. 386p [ Links ]
French EB, Gutarra L. Aley P and Elphinstone J. 1995. Culture media for Ralstonia solanacearum isolation, identification and maintenance. Fitopatologia 30:126-30. Disponible en línea: https://betuco.be/Potato/Bacterial%20wilt%20-%20culture%20media%20of%20Ralstonia20Solanacearum.pdf [ Links ]
Ganen STS, Nietsche S, Pereira MCT, Reis ST, Xavier AA, Santos TM and Fernandes TP. 2009. Microbial contamination in explants of banana cultivars ‘galil 18’ and ‘tropical’. Acta Horticulturae 829:341-344. http://dx.doi.org/10.17660/ActaHortic.2009.829.53 [ Links ]
Gillings M, Fahy P and Davies C. 1993. Restriction analysis of an amplified polygalacturonase gene fragment differentiates strains of the phytopathogenic bacterium Pseudomonas solanacearum. Letters in Applied Microbiology 17:44-48. http://dx.doi.org/10.1111/j.1472-765X.1993.tb01432.x [ Links ]
Hayward AC. 1964. Characteristics of Pseudomonas solanacearum. Journal of Applied Bacteriology 27: 265-277. http://dx.doi.org/10.1111/j.1365-2672.1964.tb04912.x. [ Links ]
Hong JC and Norman DJ, Reed DL, Momol MT, Jones JB, 2012. Diversity among Ralstonia solanacearum strains isolated from the southeastern United States. Phytopathology 102: 924-936. http://dx.doi.org/10.1094/PHYTO-12-11-0342 [ Links ]
Kalpage M and De Costa D. 2015. Isolation of bacteriophages and determination of their efficiency in controlling Ralstonia solanacearum causing bacterial wilt of tomato. Tropical Agricultural Research 26:140-151. http://dx.doi.org/10.4038/tar.v26i1.8079 [ Links ]
Kelman A. 1954. The relationship of pathogenicity in P. solanacearum to colony appearance on a tetrazolium medium. Phytopathology 44:693-695. Disponible en línea: http://garfield.library.upenn.edu/classics1983/A1983QM45400001.pdf [ Links ]
King EO, Ward MK and Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. Journal of Laboratory and Clinical Medicine 44:301-307. [ Links ]
Korus KA. 2007. Evaluating commercially available diagnostic tests for the detection of Clavibacter michiganensis subsp. nebraskensis, cause of Goss’s bacterial wilt and leaf blight in corn. Tesis de Maestría. Universidad de Nebraska. 73p. Disponible en línea: http://digitalcommons.unl.edu/cgi/viewcontent.cgiarticle=1025&context=agronhortdiss [ Links ]
Lamichhane JR and Varvaro L. 2013. A new medium for the detection of fluorescent pigment production by pseudomonads. Plant Pathology 62:624-632. http://dx.doi.org/10.1111/j.1365-3059.2012.02670.x [ Links ]
Lee Y-A and Chin N-K. 2003. A novel Insertion sequence, ISRso19, isolated from Ralstonia solanacearum and its application to race differentiation. Plant Pathology Bulletin 12:57-64. http://dx.doi.org/140.112.183.1/cpps/pdf/12-1/12-1-7 [ Links ]
Li Y, Feng J, Liu H, Wang L, Hsiang T, Li X and Huang J. 2016. Genetic Diversity and Pathogenicity of Ralstonia solanacearum Causing Tobacco Bacterial Wilt in China. Plant Disease 100:1288-1296. http://dx.doi.org/10.1094/PDIS-04-15-0384-RE [ Links ]
Naranjo Feliciano E y Martínez Zubiaur Y. 2013. Avances en el diagnóstico de la marchitez bacteriana (Ralstonia solanacearum): situación actual y perspectivas en Cuba. Revista de Protección Vegetal 28: 160-170. Disponible en línea: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S1010-27522013000300001 [ Links ]
Nasim B. 2011. Biological diversity of Ralstonia solanacearum strains causing Bacterrial Wilt of solanaceous crops in Pakistan. Arid Agriculture University, Rawalpindi. 153p. Disponible en línea: http://eprints.hec.gov.pk/7587/1/1063S.htm [ Links ]
Norman DJ, Huang Q, Yuen JMF, Mangravita-Novo A and Byrne D. 2009. Susceptibility of Geranium Cultivars to Ralstonia solanacearum. HortScience 44:1504-1508. Disponible en línea: http://hortsci.ashspublications.org/content/44/5/1504.full [ Links ]
Opina N, Tavner F, Holloway G, Wang JF, Li TH, Maghirand R, Fegan M, Hayward AC, Krishnapillai V, Hong WF, Holloway BW and Timmis JN. 1997. A novel method for development of species and strain - specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formely Pseudomonas solanacearum). Asia Pacific Journal of Molecular Biology and Biotechnology 5:19-33. Disponible en línea: https://www.researchgate.net/publication/37625749 [ Links ]
Peeters N, Guidot A, Vailleau F and Valls M. 2013. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Molecular Plant Pathology 14:651-662. http://dx.doi.org/10.1111/mpp.12038 [ Links ]
Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B and Allen C. 2016. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17:1-11. http://dx.doi.org/10.1186/s12864-016-2413-z [ Links ]
Rajeshwari N, Shylaja MD, Krishnappa M, Shetty HS, Mortensen CN and Mathur SB. 1998. Development of ELISA for the detection of Ralstonia solanacearum in tomato: its application in seed health testing. World Journal of Microbiology and Biotechnology 14: 697-704. http://dx.doi.org/10.1023/A:1008892400077 [ Links ]
Ramesh, R, Achari GA and Gaitonde S. 2014. Genetic diversity of Ralstonia solanacearum infecting solanaceous vegetables from India reveals the existence of unknown or newer sequevars of Phylotype I strains. European Journal of Plant Pathology 140:543-562, http://dx.doi:10.1007/s10658-014-0487-5 [ Links ]
Raymundo AK, Aves-Ilagan Y and Denny TP. 1998. Analysis of genetic variation of a population of banana infecting strains of Ralstonia solanacearum. Pp: 56-60. In: Prior P, Allen C and Elphinstone J (eds.). Bacterial wilt disease: Molecular and ecological aspects. Paris: INRA Editions. [ Links ]
Ruiz-García N, Mora-Aguilera G, Rivas-Valencia P, Góngora-Canul C, Loeza-Kuk E, Ochoa-Martínez D, Ramírez-Valverde G, Gutiérrez-Espinosa MA and Álvarez-Ramos R. 2009. Sensibilidad de inmunoimpresión-ELISA y DAS-ELISA en el diagnóstico y muestreo del virus de la tristeza de los cítricos en huertos comerciales de Tamaulipas, México. Revista Chapingo Serie Horticultura 15:41-47. Disponible en línea: http://www.scielo.org.mx/scielo.php?script=sci_arttext&pid=S1027-152X2009000100007 [ Links ]
Safni I, Cleenwerck I, De Vos P, Fegan M, Sly L and Kappler U. 2014. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. International Journal of Systematic and Evolutionary Microbiology 64:3087-3103. http://dx.doi.org/ 10.1099/ijs.0.066712-0 [ Links ]
Sambrook J and Russell DW. 2001. Molecular cloning: a laboratory manual. Third edition. Cold Spring Harbor Laboratory Press. United States of America. 2100p. [ Links ]
Sánchez-Pérez A, Mejia L, Fegan M and Allen C. 2008. Diversity and distribution of Ralstonia solanacearum strains in Guatemala and rare occurrence of tomato fruit infection. Plant Pathology 57:320-331. http://dx.doi.org/10.1111/j.1365-3059.2007.01769.x [ Links ]
SENASICA, 2015. Campaña contra el Moko de platano. Informe mensual No. 12 (Diciembre 2015). 6p. Disponible en línea: Disponible en línea: http://cesavenay.org.mx/wp-content/uploads/Informes-Fisicos-Financieros/2015/12-IFF-DICIEMBRE-MOKO.PDF [ Links ]
SENASICA, 2016. Moko del plátano. Ralstonia solanacearum raza 2 Smith. Ficha técnica No. 3. 24p. Disponible en línea: http://www.gob.mx/cms/uploads/attachment/file/130955/Ficha_Técnica.pdf [ Links ]
SENASICA, 2017. Campaña fitosanitaria Moko del plátano. Disponible en línea: http://www.gob.mx/senasica/acciones-y-programas/plaga-Moko-del-platano. Fecha de consulta 04 de enero del 2017. [ Links ]
SIAP. Servicio de información Agroalimentaría y Pesquera. 2017. Disponible en línea: Disponible en línea: http://www.gob.mx/siap/acciones-y-programas/produccion-agricola-33119 . (consulta, enero 2017). [ Links ]
Sequeira L. 1998. Bacterial wilt: the missing element in international banana improvement programs. Pp 6-14. In: Prior P, Ass CA, Elphinstone J (eds). Bacterial Wilt Disease. Springer-Verlag, Berlin, Heidelberg. 145p. [ Links ]
Smith EF. 1896. A bacterial disease of tomato, pepper, eggplant, and Irish potato (Bacillus solanacearum nov. sp.). U. S. Dept. Agric. Div. Vegetable Physiol. Pathol. Bulletin 12:1-28. Disponible en línea: https://archive.org/stream/bacterialdisease12smit#page/n1/mode/2up [ Links ]
Souza SA, Xavier AA, Costa MR, Cardoso AMS, Pereira MCT and Nietsche S. 2013. Endophytic bacterial diversity in banana ‘Prata Anã’ (Musa spp.) roots. Genetics and Molecular Biology 36:252-264. http://dx.doi.org/10.1590/S1415-47572013000200016 [ Links ]
Thera AT, 2007. Bacterial wilt management: a prerequisite for a potato seed certification program in Mali. Montana State University-Bozeman, College of Agriculture. 142p. Disponible en línea: http://scholarworks.montana.edu/xmlui/bitstream/handle/1/2415/TheraA1207.pdf?sequence=1&isAllowed=y [ Links ]
Thomas P, Swarna GK, Roy PK and Patil P. 2008. Identification of culturable and originally non-culturable endophytic bacteria isolated from shoot tip cultures of banana cv. Grand Naine. Plant Cell, Tissue and Organ Culture 93:55-63. http://dx.doi.org/10.1007/s11240-008-9341-9 [ Links ]
Wicker E, Grassart L, Coranson-Beaudu R, Mian D, Guilbaud C, Fegan M and Prior P. 2007. Ralstonia solanacearum strains from Martinique (French West Indies) exhibiting a new pathogenic potential. Applied and Environmental Microbiology 73:6790-6801. http://dx.doi:10.1128/aem.00841-07 [ Links ]
Wicker E, Grassart L, Coranson-Beaudu R, Mian D and Prior P. 2009. Epidemiological evidence for the emergence of a new pathogenic variant of Ralstonia solanacearum in Martinique (French West Indies). Plant Pathology 58:853-861. http://dx.doi:10.1111/j.1365-3059.2009.02098.x [ Links ]
Workman D. 2017. Bananas Exports by Country. Disponible en línea: Disponible en línea: http://www.worldstopexports.com/bananas-exports-country/ . Fecha de consulta 24 de julio del 2017. [ Links ]
Acknowledgements
This research was carried out with the authorization of the DGSV, SENASICA, Mexico. We would like to thank all the support given for this project by Dr. Francisco Javier Trujillo-Arriaga, Director of the DGSV, Dr. José Abel López-Buenfil, M.C. Oscar Morales-Galván and the staff of the Bacteriology Laboratory for Technical training of José Abraham Obrador Sánchez (JAOS) in the National Plant Health Reference Center (Centro Nacional de Referencia Fitosanitaria - CNRF). The authors would also like to thank the Tabasco Local Plant Health Committee (Comité Local de Sanidad Vegetal de Tabasco - CESVETAB) for the sampling of banana plants with Moko symptoms, and the Phytosanitary Epidemiological Surveillance Program (Programa de Vigilancia Epidemiológica Fitosanitaria - PVEF) for the shipping of samples to our lab. This research was funded by the National Science and Technology Council (Consejo Nacional de Ciencia y Tecnología - CONACyT), with Project 60246 and scholarship 344769 to JAOS, and partially supported by CONACYT project 269833.
Received: May 05, 2017; Accepted: July 24, 2017











 text in
text in 


