Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.35 no.3 Texcoco sep. 2017
https://doi.org/10.18781/r.mex.fit.1612-7
Scientific articles
Genetic diversity of Hemileia vastatrix of two coffee producing areas in Peru
1Universidad Nacional Agraria La Molina, Av. La Molina S/N, Lima, Perú, C.P. 12056.
The genetic diversity of the coffee pathogen Hemileia vastatrix was analyzed by sequencing the internal transcribed spacer (ITS) of ribosomal DNA. The analysis was carried out using samples of rust pustules from 12 properties of two coffee areas in Peru, Quillabamba (Cusco) and Villa Rica (Pasco). Haplotype diversity indexes were found and the similarities among the populations analyzed were determined using a haplotype network. From the analysis, it was determined that both producing regions presented high values of genetic diversity, but Quillabamaba harbored the highest haplotype diversity of H. vastatrix (Hd = 0.977 +/- 0.012). In addition, the haplotype network showed the structure of H. vastatrix populations for these two areas, which behave as a large undifferentiated population with the presence of ancestral haplotypes from which new variants of the fungus were generated. We also analyzed the sequences of the ITS region of H. vastatrix collected in Peru and those reported in the GenBank, but we did not find any distribution of the populations by geographic region; it was also suggested that the populations of Peruvian rust presented haplotypes similar to those from Colombia and races II and XXII.
Key words: Yellow rust; ITS; Coffea spp; haplotypes
Se analizó la diversidad genética del patógeno del café Hemileia vastatrix, a través de secuenciación de los espaciadores internos transcribibles del ADN ribosomal (ITS). El análisis se realizó en muestras de roya de 12 predios de dos zonas cafetaleras del Perú, Quillabamba (Cusco) y Villa Rica (Pasco). Se hallaron los índices de diversidad haplotípica y nucleotídica. Además, se determinaron las semejanzas entre las poblaciones analizadas por medio de una red de haplotipos. Del análisis, se determinó que ambas regiones productoras presentan valores altos de diversidad genética; sin embargo, la zona de Quillabamba albergó la mayor diversidad haplotípica de H. vastatrix (Hd = 0.977+/- 0.012). Asimismo, la red de haplotipos permitió evidenciar la estructura de las poblaciones de H. vastatrix para cada zona, las que juntas se comportan como una gran población indiferenciada con presencia de haplotipos ancestrales a partir de los cuales se fueron generando nuevas variantes del hongo. También, se analizaron las secuencias de la región ITS de H. vastatrix peruanas y las almacenadas en el GenBank, no encontrándose distribución de las poblaciones por región geográfica; además, se determinó que las poblaciones de H. vastatrix peruanas presentan haplotipos similares a los de Colombia y a las razas II y XXII.
Palabras clave: Roya amarilla; ITS; Coffea spp; haplotipos
INTRODUCTION
Coffee (Coffea spp.) is the main agricultural product exported from Peru, where it is sown on 361 671 ha that produce 222 047 tons mainly in the regions of Cajamarca, Amazonas, Junín, Cusco and San Martín (MINAGRI, 2015). Ninety five percent of Peruvian coffee comes from Coffea arabica cultivars (JNC, 2014) that are very susceptible to yellow rust, a disease caused by the biotrophic fungus Hemileia vastatrix (Berkeley, 1869).
Yellow rust of coffee was first reported in Peru in 1979 in the Department of Junín (Scheiber and Zentmyer, 1984). However, in 2013 it had the highest incidence causing 27% production losses (INEI, 2014) that mainly affected organic producers. Since then, yellow rust has spread to most of Peru’s main coffee areas, including Villa Rica (Pasco Region) and Quillabamba (Cusco Region).
Yellow rust is considered the most damaging coffee disease worldwide and may cause up to 40% production losses (Rivillas et al., 2011). Two approaches have been proposed to prevent losses caused by this disease. The first is fungicide use, which is expensive and potentially harmful to the environment; the second is to develop improved varieties. However, for the latter it is necessary to know the sources of resistance and the pathogen’s diversity.
The mechanisms that promote the emergence of new virulent strains of H. vastatrix are still unknown, and it has been suggested that mutations are the main cause of increased fungal diversity (Varzea and Marques, 2005). However, there is evidence of a new type of hidden sexual reproduction within asexual spores (cryptosexuality) that could explain the origin of new races (Carvalho et al., 2011). It is also known that the high rate at which new races develop -an issue that has attracted the interest of genetic improvement programs- has affected the way new coffee varieties are generated, because the fungus develops the ability to infect plants that were originally resistant (Alvarado and Moreno, 2005).
Due to the ability of the fungus to quickly adapt to new coffee cultivars, H. vastatrix has been considered a pathogen with high evolutionary potential whose variation has been studied since 1930 (Rodrigues et al., 1975). Studies have reported the presence of race II in almost all cases. For this reason, it has been suggested that race II is likely the initial inoculum from which all the new races found today were naturally produced (Zambolim et al., 2005). These initial studies were conducted through readings of the phenotypic expressions of a series of clones of differentiated coffee trees from the Centro de Invesgação das Ferrugens do Cafeeiro (CIFC) in Portugal; by 2005, 45 races of H. vastatrix had been identified (Varzea and Marques, 2005). However, since these sets of differentiated coffee trees were not enough to characterize new virulent races of the fungus, new techniques (such as molecular markers) (Nandris et al., 1998) and ITS sequencing of ribosomal DNA (Cristancho et al., 2007) were used to determine their variability. Although it was not possible to identify races by using these molecular techniques, they did allow determining the pathogen’s genetic diversity. Furthermore, ITS sequences have been used extensively to perform molecular analyses of the diversity of phytopathogenic fungi, characterize new fungal species and separate very closely related species or races of the same species (Grube and Kroken, 2000). Applying ITS sequences to H. vastatrix has provided new opportunities to use its diversity in genetic improvement programs. In Peru, where few studies have been conducted for this purpose, developing this knowledge is essential for crop management and for Peruvian coffee genetic improvement programs. For this reason, in this research, we analyzed the genetic diversity of H. vastatrix from two major coffee producing areas in Peru using sequences of the ITS region and then comparing them with the sequences reported by the GenBank for Colombia.
MATERIALS AND METHODS
Biological material
Hemileia vastatrix spores were collected in five plots in the Quillabamba coffee producing area (Cusco Region) and in seven plots in the Villa Rica coffee producing area (Pasco Region) (Table 1). These areas are geographically distant and considered to be among the highest coffee-producing regions in Peru (Figure 1).
In each plot, we gathered 25 leaves from coffee trees of the Typica and Caturra varieties that are infected with H. vastatrix and extracted spores. To obtain enough DNA concentration, samples from one or two plots were grouped to form a compound sample (Table 1).
Table 1 Geographic information of the plots where samples of pustules of 25 leaves of Hemileia vastatrix were collected in the two Peruvian coffee production areas; it includes keys of the compounded samples formed.

Analysis of genetic diversity in Quillabamba and Villa Rica
DNA extraction
DNA was extracted using the protocol described by Cristancho et al. (2007) with modifications: uredospores were placed in 1.5 tubes to which 150 μL of lysis buffer was added (50 mM of tris HCl pH 7,5; 50 mM of EDTA pH 8,0; 3% of SDS; 1% of b mercaptoethanol); the uredospores were then ground for 30 to 45 minutes using a polypropylene pestle. The quality was determined by electrophoresis in 1% agarose gels stained with GelRed (Biotium®). Finally, DNA concentration was determined by spectrophotometry.
PCR and Purification
The ITS region was amplified using PCR and ITS1L and ITS4R universal primers (White et al., 1990); as a reference, we used the protocol described by Cristancho et al. (2007). The amplification reaction was performed in a 10 μL volume containing 2 mM of MgCl2, 0.2 mM of dNTPs, 0.4 μM of each primer, PCR 1X buffer, 1.5 units of Taq DNA polymerase and 40 ng of DNA. The amplification procedure consisted of an initial stage at 95 °C for 5 min, followed by 30 denaturation cycles at 94 °C for 1.5 min, alignment at 60 °C for 1 min and extension at 72ºC for 2 min, plus a final extension at 72ºC for 5 min. The amplification products were separated by electrophoresis in 1% agarose gels; a QIAquick gel extraction Kit® was used to purify the DNA fragment of the ITS region.
Cloning and sequencing
The purified amplified products were cloned in chemocompetent Escherichia coli (JM109) using the Pgem T-easy vector system II Kit®, following the protocol recommended by the manufacturer. Then, for each compound sample, 10 white colonies were selected and each was sown in 5 ml of LB medium with 60 μg/mL of ampicillin, and then incubated at 37 °C for 24 h. The E. coli plasmid containing the DNA fragment of interest was extracted using the protocol of the Wizard®Plus SV Minipreps DNA purification system from PROMEGA. The presence of the DNA sequence of the ITS region was confirmed through digestion with the EcoRI enzyme. The fragments were visualized in 1.5% agarose gel. The sequencing of the cloned fragments was performed by the GenBioteck company (Argentina).
Data analysis
The obtained sequences were aligned and edited using the Codon Code Aligner v 2.0.6 program (Codon Code Corporation). As a reference, we used the DNA sequences of the ITS region of H. vastatrix stored in the GenBank (Cristancho et al., 2007).
The obtained sequences were aligned using the MAFFT program implemented in the UGENE platform (Golosova et al., 2014). Nucleotide and haplotype diversities (Nei and Li, 1979) were calculated using the DNAsp program version 5 (Librado and Rozas, 2009), and to detect similarities among haplotypes a network was created using the Median Joining algorithm (MJ) implemented in the PopArt program (Bandelt et al., 1999) from which gap sites were excluded.
We also analyzed sequences of the ITS regions of Hemileia vastatrix stored in the GenBank, which correspond to H. vastatrix from Colombia, and of races II and XXII from CIFC (Cristancho et al., 2007).
RESULTS AND DISCUSSION
Analysis of the genetic diversity in Quillabamba and Villa Rica
From samples of H. vastatrix collected in plots in Quillababa (5 plots) and Villa Rica (7 plots), we obtained 106 DNA sequences of the ITS region of Hemileia vastatrix. The length of the sequences ranged between 910 pb and 918 pb.
By aligning these sequences, we obtained 921 aligned sites, of which 21 were gaps (insertions or deletions). We also found 101 polymorphic sites with 11 informative parsimonious positions (Table 2).
Table 2 Values of trameters obtained from descriptive measures of Hemileia vastatrix variability using sequences of the ITS region associated with samples from Quillabamba, Villa Rica, Colombia and CIFC.
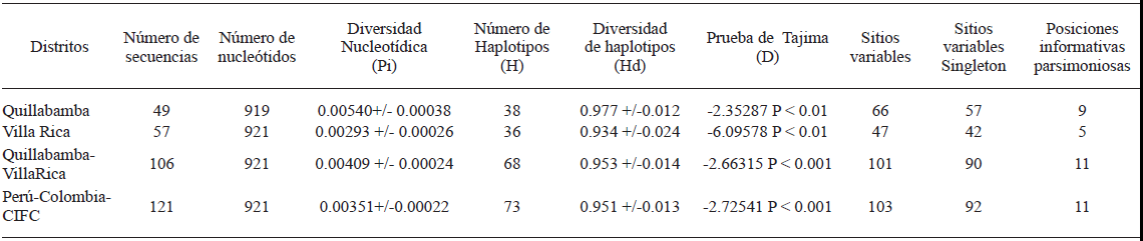
CIFC: Centro de Invesgação das Ferrugens do Cafeeiro.
In the two coffee producing areas, a total of 68 haplotypes was found with haplotype (Hd = 0.953 +/- 0.014) and nucleotide (Pi = de 0.00409 +/- 0.00024) diversity; 62 haplotypes showed unique sequences and 6 haplotypes consisted of more than two sequences (Table 3). Haplotypes Hap_4, Hap_8, Hap_36 and Hap_21 were found more often and are considered ancestors (Figure 2); they are also distributed across both coffee-producing areas, where they must have arrived at different times and became adapted to each area’s environmental conditions. Haplotypes Hap_4 and Hap_36 are found more often in Villa Rica, while Hap_8 and Hap_21 are found more often in Quillabamba. The distribution of these ancestors in the two areas is explained by the coalescence genetic theory also observed by Santana et al. (2007) in populations of the fungus in Brazil.
Table 3 Cont’d… Regional distribution of the occurrence of H. vastatrix haplotypes obtained from ITS sequences.
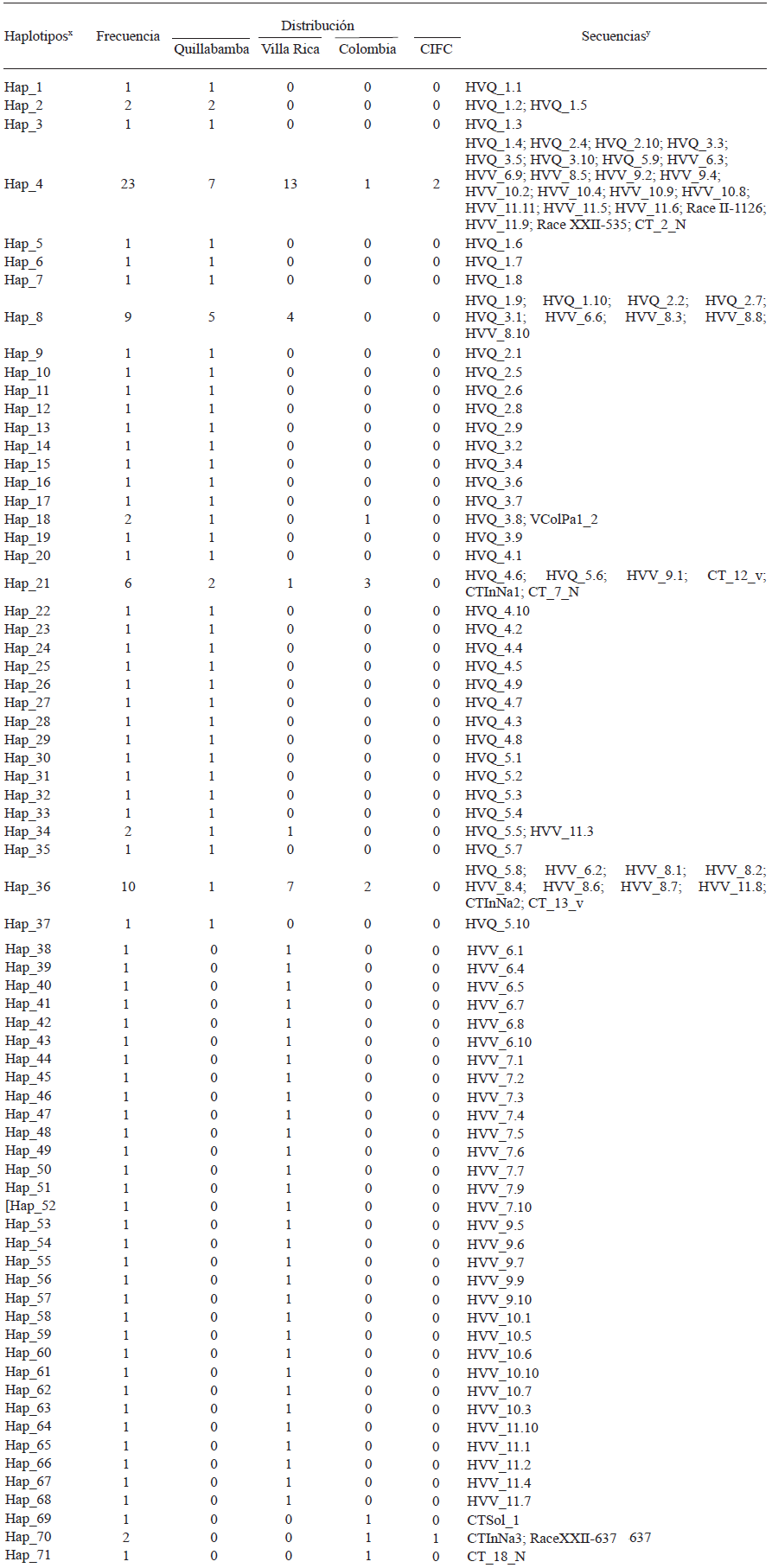
x Hap: Haplotype.
y HVQ: Hemileia vastatrix from Quillabamba; HVV: Hemileia vastatrix from Villa Rica; CT: Samples of yellow rust from Colombia; Race XXII and Race II: Samples corresponding to CIFC.
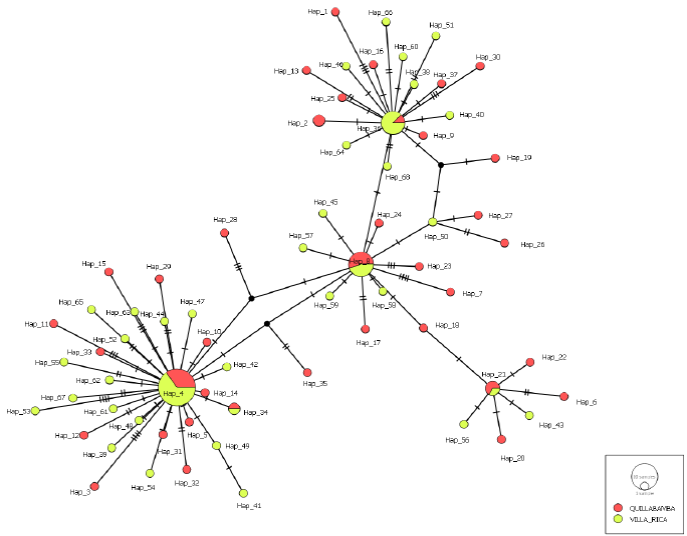
Figure 2 Haplotype network created using the Median Joinig method for 68 haplotypes detected for the ITS region of Hemileia vastatrix from Quillabamba and Villa Rica coffee production areas. Circles represent a unique haplotype; the size of the circles is proportional to the number of sequences of a haplotype; transverse lines represent the events of simple mutations, and black nodes lost or non-sampled haplotypes.
From the analysis of H. vastatrix populations from Quillabamba and Villa Rica, we concluded that the highest genetic diversity is present in the Quillabamba coffee producing area, because of its higher nucleotide and haplotype diversity values (Table 2).
The absence of exclusive nodes by geographic region in the haplotype network in Figure 2 would indicate a large and undifferentiated population of H. vastatrix that may have resulted from the flow of spores between the two areas carried by people, seed and/or infected plants. These results are similar to those obtained in Brazil by Cabral et al. (2016), who did not observe that H. vastatrix populations were structured based on their origin, host or physiological race. However, the genetic diversity was relatively lower than the diversity found in the present study. However, Maia et al. (2013) (also in Brazil) obtained results similar to those of the present study using 91 fungal isolates; they proposed that H. vastatrix acts as a large indifferentiated population with high genotypic diversity that is not structured based on its geographic origin and host. Therefore, the lack of structure of the two H. vastatrix populations by region may be explained by the fact that the fungus is spread over long distances.
Tajima’s neutrality test was used for H. vastatrix populations from Villa Rica (D = -2.50181; P < 0.001) and Quillabamba (D = -2.35287; P < 0.001), and for both areas together (D= -2.66315; P < 0.001). These values are lower than 0, which indicate that H. vastatrix populations are in process of expansion. Also, they may have originated from a few recently introduced genotypes, or may descend from race II, which, according to Scheiber and Zentmyer (1984) exists in Peru since 1979. These Tajima’s D test negative values may explain the presence of a high number of unique haplotypes in the haplotype network that are increasing the variability due to accelerated population growth. Therefore, this reflects the status of the H. vastatrix population and shows that coffee yellow rust epidemics in Peru, which started in 2013, were initially caused by a few individuals. However, it was not possible to establish the effect of the altitude above sea level on the population, because the Tajima test negative value indicates that there is no evidence of the existence of a selection process. The mechanisms that increase variability and create new H. vastatrix races are unknown. Therefore, spontaneous mutations may be the main mechanism responsible for H. vastatrix variability, especially in seasons of higher disease incidence. Another reason could be the selection pressure exerted by the environment. In this regard, Varzea and Marques (2005) believe that in Brazil the presence of resistance genes in some varieties of the host species increases the number of virulent mutants in the population. Another factor that could explain the frequent and rapid emergence of new variants is cryptosexuality (Carvalho et al., 2011), which may have an important role in creating H. vastatrix genetic variability.
Comparing H. vastatrix haplotypes from Peru to haplotypes from Colombia
In the analysis of the ITS sequences of Peruvian H. vastatrix populations, we included 15 DNA sequences from the ITS region of the fungus that are stored in the GenBank; of these, 12 sequences correspond to H. vastatrix from Colombia, and 3 from CIFC. The latter are race II and XXII haplotypes (Figure 3). The analysis found a total of 73 haplotypes, 65 of which were unique (they had no replications). Also, it was confirmed that haplotype Hap_4 contains the races II and XXII. It is frequently present and distributed across all the studied areas (7 sequences from Quillabamba, 13 from Villa Rica, 1 from Colombia and 2 from CIFC). Therefore, we may assume that the other haplotypes emerged from this ancestral haplotype, which confirms the findings of Zambolim et al. (2005) in Brazil, who suggested that the other races emerged from race II, which prevails in Latin America (Cristancho et al., 2007). It was also observed that haplotypes Hap_36 and Hap_21, which are present in Colombia, Villa Rica and Quillabamba, are nodes that appeared later. Haplotype Hap_8 is present only in Quillabamba and Villa Rica; after being introduced or emerging in Peru, this haplotype was able to better adapt to the environmental conditions of Peru’s coffee producing areas. Based on what we observed in the main nodes (in which the presence of H. vastatrix sequences from Colombia and Peru is repetitive), it may be that in Peru and Colombia the disease emerged in the same area four years apart (Scheiber and Zentmyer, 1984; Buriticá, 2010). No nodes by origin of provenance were observed in the network, which would indicate that H. vastatrix sequences from Colombia are similar to the sequences from Peru. Studies conducted in early 1970 indicate that the wind has an important role in spreading the disease. Martines et al. (1975) found uredospores with good germination viability at an altitude of more than 1000 m; it has also been suggested that the introduction and rapid spread of the fungus in South America was caused by the wind (Bowden et al., 1971). Other disease spreading means have also been mentioned, for example, the disease was introduced into Brazil through plants from Africa carried by people (migrant workers) and seed (Schieber, 1972). Therefore, anthropogenic causes may have had a very important role in bringing the fungus to Peru. Based on the analysis of H. vastatrix populations from the studied regions, three hypotheses about the origin of the 2013 yellow rust epidemic in Peru may be proposed. The first hypothesis assumes that a new H. vastatrix race derived from Race II was introduced in Peru and quickly adapted to the environmental conditions of coffee producing areas at altitudes higher than 1000 masl. Also, because the main ancestral haplotypes were found more frequently in Villa Rica, they must have arrived earlier in this area than in Quillabamba. However, in the haplotype network there is more than one main node that is considered ancestral (Hap_4, Hap_8, Hap_21 and Hap_36). The four haplotypes must have arrived in Peru progressively and in a short time. Haplotype Hap_4, that corresponds to race II, is more frequently present in both coffee producing areas and includes a CT_2N sequence from Colombia in its background.
The second hypothesis states that the H. vastatrix population evolved from race II and that a variant emerged that is able to infect high-altitude coffee trees. However, based on what we observed in the haplotype network (Figure 3), at least four variants corresponding to the ancestral haplotypes must have developed in hight altitude. However, two of those sequences of both Colombia and Peru (Hap_4 and Hap_36) may have been introduced to Colombia from Peru, but this is very unlikely because the movement of all plant materials is under government control (Cristancho et al., 2007).
The third hypothesis states that changes in weather conditions may have favored the fungus to reproduce and spread toward areas above 1000 masl. This hypothesis is supported by what is observed in the haplotype network (Figure 3), in which 4 nodes are formed that correspond to the ancestral haplotypes (Hap_4, Hap_8, Hap_21 and Hap_36) which may have existed years ago, because they are present in both coffee producing areas of Peru, and are spreading. Also, the most frequently found haplotype is Hap_4, which corresponds to race II and has been present in Peru since 1979. This hypothesis is similar to the one proposed by Cristancho et al. (2012) in Colombia as the origin of the coffee epidemic in 2008 that affected Colombian coffee producing areas above 1400 masl.
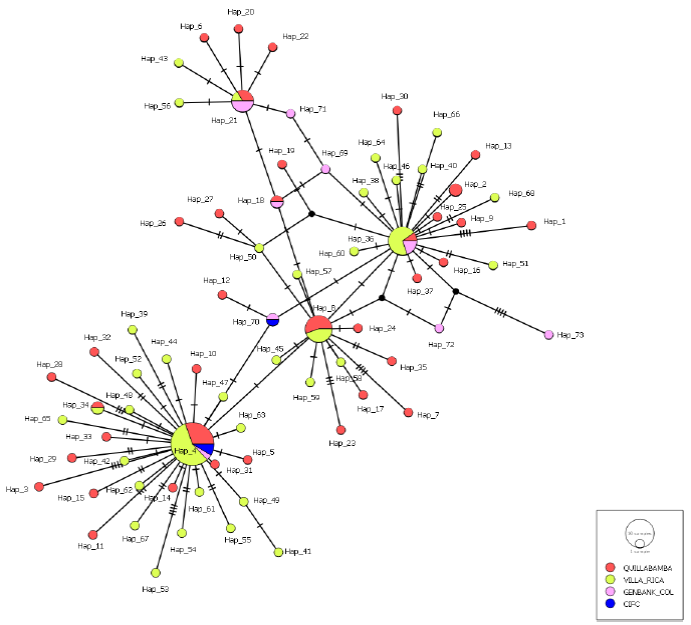
Figure 3 Haplotype network created using the Median Joinig method of 73 haplotypes detected for the ITS region of Hemileia vastatrix from Quillabamba, Villa Rica, Colombia and CIFC. Circles represent a unique haplotype; the size of the circles is proportional to the number of sequences of a haplotype; transverse lines represent the events of simple mutations, and black nodes lost or non-sampled haplotypes.
CONCLUSIONS
By analyzing Hemileia vastatrix genetic diversity using ITS region sequences, we concluded that the haplotypic diversity values for Quillabamba and Villa Rica in Peru were high (0.934 +/- 0.024 and 0.977 +/- 0.012, respectively). However, the nucleotic diversity was low (0.00293 +/- 0.00026 and 0.00540 +/- 0.00038, respectively). However, the highest H. vastatrix diversity was found in the Quillabamba coffee producing area. In both studied areas, a large non-differentiated population was found by area of provenance, and their haplotypes are similar to those from Colombia. The sequences of races II and XXII stored in the GenBank that correspond to haplotype Hap_4 are present both in Quillabamba and Villa Rica. Also, three hypotheses are proposed to explain the increase of rust intensity in Peru.
REFERENCES
Alvarado-Alvarado G and Moreno-Ruiz G. 2005. Cambio de la virulencia de Hemileia vastatrix en progenies de Caturra x Hibrido de Timor. Cenicafe 56:110-126. http://biblioteca.cenicafe.org/handle/10778/185 [ Links ]
Bandelt H, Forster P and Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16(1):37-48. http://mbe.oxfordjournals.org/content/16/1/37.full.pdf+html [ Links ]
Berkeley MJ. 1869. [Untitled]. Gardener’s Chronicle 45: 1157. [ Links ]
Bowden J, Gregory PH and Johnson CG. 1971. Possible wind transport of coffee leaf rust across the Atlantic Ocean. Nature 224:500-501. https://doi.org/10.1038/229500b0 [ Links ]
Buriticá CP. 2010. La Roya del Cafeto en Colombia: Realizaciones de Impacto Nacional e Internacional en el Siglo XX. Revista Facultad Nacional de Agronomía. Medellín, 63(1): 5285-5292. http://www.redalyc.org/articulo.oa?id=179914617007 [ Links ]
Cabral PGC, Maciel-Zambolim E, Oliveira SAS, Caixeta ET and Zambolim L. 2016. Genetic diversity and structure of Hemileia vastatrix populations on Coffea spp. Plant Pathology 65:196-204. https://doi.org/10.1111/ppa.12411 [ Links ]
Carvalho CR, Fernandes RC, Carvalho GMA, Barreto RW and Evans HC. 2011. Cryptosexuality and the Genetic Diversity Paradox in Coffee Rust, Hemileia vastatrix. Plos one 6(11): 1-7. https://doi.org/10.1371/journal.pone.0026387 [ Links ]
Cristancho AM, Escobar OC and Ocampo JD. 2007. Evolución de razas de H. vastatrix en Colombia. Cenicafé 58(4): 340-359. http://biblioteca.cenicafe.org/bitstream/10778/178/1/arc058%2804%29340-359.pdf [ Links ]
Cristancho MA, Rozo Y, Escobar C, Rivillas CA and Gaitán AL. 2012. Outbreak of coffee leaf rust (Hemileia vastatrix) in Colombia. New Disease Reports, 25, 19. https://doi.org/10.5197/j.2044-0588.2012.025.019 [ Links ]
Golosova O, Henderson R, Vaskin Y, Gabrielian A, Grekhov G, Nagarajan V, Oler AJ, Quiñones M, Hurt D, Fursov M and Huyen Y. 2014. Unipro UGENE NGS pipelines and components for variant calling, RNA-seq and ChIP-seq data analyses. PeerJ 2: 1-15. http://dx.doi.org/10.7717/peerj.644 [ Links ]
Grube M and Kroken S. 2000. Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycological Research 104: 1284-1294. https://doi.org/10.1017/s0953756200003476 [ Links ]
INEI, Instituto Nacional de Estadística e Informática. 2014. Compendio estadístico del Perú. https://www.inei.gob.pe/media/MenuRecursivo/publicaciones_digitales/Est/Lib1173/cap12/cap12.pdf [ Links ]
JNC, Junta Nacional del Café. 2014. Plan Nacional de Renovación de cafetales. http://juntadelcafe.org.pe/publicaciones/documento-plan-nacional-de-renovacion-de-cafetales [ Links ]
Librado P and Rozas J. 2009. DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451-1452. http://diposit.ub.edu/dspace/bitstream/2445/53352/1/568571.pdf [ Links ]
Maia TA, Maciel-Zambolim E, Caixeta ET, Mizubuti ESG and Zambolim L. 2013. The population structure of Hemileia vastatrix in Brazil inferred from AFLP. Australasian Plant Pathology 42(5): 533-542. http://dx.doi.org/10.1007/s13313-013-0213-3 [ Links ]
Martínez JA, Palazzo DA, Karazawa M, Monteiro MVM e Reu NRN. 1975. Presença de esporos de Hemileia vastatrix Berk et Br. agente causal de ferrugem do cafeeiro, em diferentes altitudes nas principais areas cafeeiras dos estados de Sao Paulo e Parana, Brasil. O Biológico 41(3): 77-88. [ Links ]
MINAGRI, Ministerio de Agricultura y Riego. 2015. Síntesis agroeconómica del café. http://repositorio.minagri.gob.pe/handle/MINAGRI/51 [ Links ]
Nandris D, Kohler F, Fernandez D, Lashermes P, Rodrigues Jr. CJ and Pellegrini PF. 1998. Coffee pathosystems modelling: 2. Assessment pathogen biodiversities. In: Proceedings of the 7th International Congress of Plant Pathology , Edinburgh, Scotland. Abstract 2.2.119. [ Links ]
Nei M and Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences, 76(10):5269-5273. http://dx.doi.org/10.1073/pnas.76.10.5269 [ Links ]
Rivillas CA, Serna CA, Cristancho MA and Gaitán A. 2011. La roya del cafeto en Colombia: Impacto, Manejo y Costos de Control. Cenicafé 36:1-51. http://biblioteca.cenicafe.org/bitstream/10778/594/1/036.pdf [ Links ]
Rodriguez CJ Jr, Bettencourt AJ and Rijo L. 1975. Races of pathogen and resistance to coffee rust. Ann Rev Phytopathology 13:49-70. http://www.annualreviews.org/doi/pdf/10.1146/annurev.py.13.090175.000405 [ Links ]
Santana MF, Zambolim EM, Oliveira LO, Caixeta ET e Zambolim L. 2007. Análise molecular do rDNA de Hemileia vastatrix. SIMPÓSIO DE PESQUISA DOS CAFÉS DO BRASIL, 5, Águas de Lindóia, SP. Anais. Brasília, DF: Embrapa Café. file:///C:/Users/Cliente%205/Downloads/Analise-molecular.pdf [ Links ]
Schieber E. 1972. Economic impact of coffee rust in Latin America. Annual Review of Phytopathology 10:491-510. http://dx.doi.org/10.1146/annurev.py.10.090172.002423 [ Links ]
Schieber E and Zentmyer GA. 1984. Coffee rust in the Western Hemisphere. Plant Disease 68(2):89-93. https://www.apsnet.org/publications/plantdisease/backissues/Documents/1984Articles/PlantDisease68n02_89.pdf [ Links ]
Varzea V and Marques DV. 2005. Population variability of Hemileia vastatrix vs Coffee durable resistance. In Zambolim L Zambolim E Várzea VMP Eds Durable resistance to coffee leaf rust.Universidade Federal de Viçosa, Viçosa Brasil. 53-74p. [ Links ]
White TJ, Bruns T, Lee S and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Inoculum 64(1):1-9. http://dx.doi.org/10.1016/B978-0-12-372180-8.50042-1 [ Links ]
Zambolim L, Zambolim EM e Várzea VMP. 2005. Durable resistance to coffee leaf rust. Universidade Federal de Viçosa, Vicosa Brasil. 450p. [ Links ]
Acknowledgements
Received: December 15, 2016; Accepted: June 26, 2017











 texto en
texto en 



