Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.35 no.3 Texcoco sep. 2017
https://doi.org/10.18781/r.mex.fit.1611-3
Scientific articles
Identification of mucoralean fungi causing soft rot in papaya (Carica papaya L.) fruit in Mexico
1Centro de Investigación en Alimentación y Desarrollo, A.C. Unidad Culiacán. Km 5.5 Carretera Culiacán-Eldorado, Campo El Diez, Culiacán, Sinaloa, México. CP. 80110.
México is one of the main papaya producers worldwide; however, yield is affected by fungal diseases such as the fruit soft rot, which causes preharvest and postharvest losses. Although it is a common disease, information related to the identification of the causal agents is scarce. The objective in this study was to identify by morphological and molecular techniques the species of mucoralean fungi responsibles of papaya soft rot. Diseased fruits were collected during May-October 2014 in production regions in Colima, Veracruz, and Oaxaca. Mucoralean fungi were isolated, their pathogenicity was determined by the Koch's postulates and fungal structures were registered. The molecular characterization was conducted by analyzing the ITS and 28S (LSU) ribosomal regions. The identification was confirmed by comparison with sequences deposited in the Genbank and by phylogenetic analysis. The strains isolated in this study were placed in monophyletic clades supporting to Gilbertella persicaria detected in the three states sampled, Mucor irregularis in Veracruz, and Rhizopus oryzae in Oaxaca as the causal agents of papaya soft rot. This is the first report of M. irregularis and R. oryzae affecting papaya fruit in Mexico. Although G. persicaria has been reported in Colima State, this study shows its presence in Oaxaca and Veracruz States.
Key words: Gilbertella persicaria; Mucor irregularis; Rhizopus oryzae; sporangia
México es uno de los principales productores de papaya a nivel mundial; sin embargo, la producción es afectada por enfermedades fungosas, siendo la pudrición blanda de frutos una causa de pérdidas en precosecha y en poscosecha. La enfermedad es común; sin embargo, la información acerca de los agentes causales es escasa. El objetivo de este trabajo fue identificar a los hongos mucorales causantes de pudrición blanda mediante caracterización morfológica y molecular. Se recolectaron frutos enfermos durante el periodo mayooctubre de 2014 en Colima, Veracruz y Oaxaca. Se aislaron hongos mucorales, se determinó la patogenicidad con los postulados de Koch y se registraron datos de estructuras fúngicas. La caracterización molecular se realizó mediante análisis de las regiones ITS1-5.8S-ITS2 y 28S (LSU) ribosomal. La identificación de las especies se confirmó por comparación con secuencias del Genbank y análisis filogenético. Las cepas aisladas en este estudio se ubicaron en clados monofiléticos soportando a las especies Gilbertella persicaria que se encontró en los tres estados muestreados, Mucor irregularis en Veracruz y Rhizopus oryzae en Oaxaca, como los agentes causales de pudrición blanda en papaya. Este es el primer reporte de M. irregularis y R. oryzae afectando frutos de papaya en México. Aunque G. persicaria ya se ha reportado causando enfermedad en Colima, aquí se demuestra su distribución en Oaxaca y Veracruz.
Palabras clave: Gilbertella persicaria; Mucor irregularis; Rhizopus oryzae; esporangios
INTRODUCTION
Every year, Mexico produces approximately 764,514 t of papaya (Carica papaya L.) (FAO, 2014). Due to its production volume and creation of income, it is important in the national and international markets; however, papaya fruits are susceptible to different microorganisms, some of the most important of which are fungi, which cause losses of 10 up to 50% due to quality losses (Morton, 1987; Suárez-Quiroz et al., 2013). Some of the mucoralean fungi reported as important plant pathogens include Gilbertella persicaria, Mucor spp., and Rhizopus spp. (Hyde et al., 2014), included in the families Choanephoraceae, Mucoraceae, and Rhizopodaceae, respectively, which cause soft rot in fruits such as pears, apples, and peaches, and vegetables such as tomatoes. They spread quickly and have been widely reported in areas with tropical and subtropical climates (Michailides and Spotts, 1990).
Mucorales are mainly saprophytic fungi that live on the soil and decomposing plants. Mucor spp. are considered a polyphyletic group, and it is therefore necessary to carry out studies in several conserved regions of the genome for its correct identification (Hyde et al., 2014; Walther et al., 2013). Gilbertella persicaria is the only species in its genus (Benny, 1991; Walther et al., 2013; Hoffmann et al., 2013) and it shows a high degree of polymorphism in terms of colonial morphology and mycelial development rate (Papp et al., 2001). Within Rhizopus, the species that stand out are R. oryzae (syn. R. arrhizus) and R. stolonifer due to the large number of cases reported (Hyde et al., 2014).
In Mexico, mucoralean fungi cause soft rot in papaya fruits in preharvest and postharvest, and therefore affect fruit yield and quality in yet uncalculated percentages. Recently, G. persicaria was reported to be affecting fruits in the state of Colima (Cruz-Lachica et al., 2016); however, the identification studies of these species are scarce and are based on the morphological characteristics of the microorganisms and in symptoms of the disease (Suárez-Quiroz et al., 2013), which, in these species, is highly susceptible to errors (Ginting et al., 1996). The aim of this study was to identify the species of mucoralean fungi that cause soft rot in papaya fruits by an identification process that included morphological and molecular characterization.
MATERIALS AND METHODS
Sample collection
The collection of fruits with symptoms of soft rot (Figure 1A) was carried out based on its incidence on cv. Maradol papaya orchards during different sample collections in 2014: two in Tecomán, Colima in May and August in the orchard El Trébol (18° 53’ 04.9” N, 103° 56’ 33.7” O), one in October in Oaxaca in the town of San José Río Verde Jamiltepec (16° 08’ 12’’ N, 97° 45’ 11’’ O), and two more carried out in October in Veracruz, in the El Diamante orchard in Cotaxtla (18°52’25’’ N, 96° 12’ 2’’ O) and in Tlalixcoyan (21° 50’ 3’’ N, 96° 10’ 35’’ O).

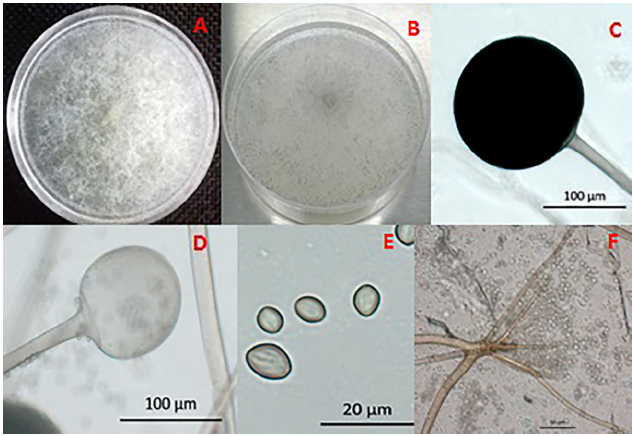
Figure 1 Pathogenicity of mucoralean strains in Maradol papaya fruits. (A) Fruit with signs and symptoms in field, (B and G) Control fruit sprayed with distilled water without symptoms, (C) Fruit with dark-brown colored morphotype 1 Gilbertella persicaria sporangia, (D) Fruit with gray to black-colored morphotype 2 Gilbertella persicaria sporangia, (E) Fruit with light brown-colored Mucor irregularis sporangia, (F) Fruit with gray to brown colored Rhizopus oryzae sporangia, and (H) Cross section of a fruit with visible internal soft rot and detachment of tissues due to enzymatic action.
Isolation, purification, and conservation of mucoralean fungi
For each fruit with symptoms, the area with lesions was divided into quadrants and a section was taken from the edge, (approximately 0.5 cm of tissue from each section) and were washed with ethyl alcohol at 70% for 1 min, followed by a later wash with distilled water. They were then placed in plates with PDA (Potato Dextrose Agar, Bioxon) and incubated at 27 °C. They were purified by transferring hyphal tips onto new PDA plates and monosporic cultures were then obtained (Chukwuka et al., 2010; Suárez-Quiroz et al., 2013). The isolates were preserved in glass tubes with sterile distilled water, and for this 20 discs of 6 mm diameter of mycelium in active growth were placed in a PDA medium from 5-day old cultures. The lid was sealed with parafilm, and they were placed at 4 °C (Ryan et al., 2012).
Pathogenicity test
Koch's postulates for selected isolates were carried out on healthy cv. Maradol papaya fruits in a comercially ripe state; initially, the fruits were disinfested with ethyl alcohol at 70% and then washed with distilled water. Each isolate was inoculated in 3 fruits, selecting three inoculation sites for each one, where incisions were made using a sterile needle and they were inoculated by spraying a sporangiospore solution at a concentration of 5 x 105 sporangiospores/mL. In addition, 3 healthy fruits were included, all free of lesions and sprayed with the same sporangiospore solution and 3 control fruits, on which lesions were made and which were sprayed with sterile distilled water only. All fruits were placed in polyethylene bags (three fruits per bag), with paper towels dampened with sterile distilled water to produce a relative humidity of about 80%, and they were incubated at 25 °C for 5 days. Finally, the fungus was reisolated from the lesions and the morpological characteristics were confirmed with the fungus inoculated at the beginning of the experiment (Beales, 2012).
Morphological Characterization
The macroscopic and microscopic cultural characteristics of the isolates grown in PDA were recorded. For this purpose, preparations were made and stained with lactophenol blue-cotton, and observed under an optic microscope (Carl Zeiss Imager A2, Germany). The appendages of sporangiospores and sporangial spines were observed using a differential interference contrast microscope (Leica DMI 6000 B, Alemania) connected to a camera (Leica DFC450C). The measurement of 100 morphological structures was recorded, and included the diameter and branching pattern of sporangiophores, columellae and chlamydospores, size and presence/absence of appendages, as well as the type of mycellium (Campbell et al., 2013).
Mycelial development rate
In the center of petri dishes, 90 mm in diameter and containing PDA, a 6 mm disc was placed, taken from the edge of fungal cultures of the isolates grown for 3 days in PDA, and they were incubated at 4, 10, 15, 25, and 40°C. The diameter of the culture was recorded on a daily basis until it covered the entire plate. Later, a relation was made between the diameter and the number of days of the test (Michailides, 1991).
Molecular Characterization
Extraction of DNA, amplification with PCR and sequencing. Each isolate’s mycelium was produced in a PDA medium for two days at 27 °C; later, it was collected with a sterilized slide and the genomic DNA was extracted according to the CTAB (cetyltrimethylammonium bromide) method, following the procedure described by Voigt et al. (1999) with slight modifications: briefly, 30 mg of mycelia were placed in 1.5 mL microcentrifuge tubes and 700 μL of CTAB buffer solution were added [100 mM Tris-Cl (pH 8.0), 1.4 M NaCl, 25 mM of EDTA, 2% CTAB], it was macerated and placed in a vortex for 10 s. Next, 700 μL of choloform-isoamylic alcohol was added (24:1) (Sigma-Aldrich) per tube; it was vortexed for 10 s and centrifuged at 12,300 x g for 10 min. A 500 μL portion of the top section was recovered and placed in a new tube; the same amount of isopropanol (Sigma-Aldrich) was added at -20 °C, and mixed softly. Later, it was centrifuged again at 12,300 x g for 2 min and the supernatant was discarded. The DNA pellet was washed with 500 μL of ethanol at 70%, centrifuged at 12,300 x g for 2 min. The ethanol was discarded and allowed to evaporate completely. Finally, the pellet was resuspended in 200 μL of Tris EDTA buffer solution [10 mM Tris-Cl (pH 8.0), 1 mM EDTA (pH 8.0)].
Amplification was carried out in accordance with Walther et al. (2013), where ribosomal DNA, including the complete region of ITS1-5.8S-ITS2 and the region D1/D2 of the subunit 28S (LSU) were amplified with the pair of oligonucleotides V9G (5’TTACGTCCCTGCCCTTTGTA3’) (de Hoog y van den Ende, 1998), and LR3 (5’-GGTCCGTGTTTCAAGAC3’) (Vilgalys and Hester, 1990). The mixture of the PCR reaction (25 μl), included 20 ng of ADN, 0.4 μM of each oligonucleotide, 0.185 mM of each deoxynucleotide triphosphate (dATP, dTTP, dGTP, dCTP), 5X reaction buffer, 1.5 mM of MgCl2 and 0.8 U of the enzyme Taq ADN polymerase using the kit GoTaq® PCR Core Systems (Promega, USA). The amplification reaction was carried out in a BioRad T100™ (Singapore) thermocycler, with the following cycles: an initial step of denaturalization for 5 min at 94 °C, followed by 35 one minute cycles at 94°C, 1 min at 53 °C, and 2 min at 72°C, with a final extension of 7 min at 72 °C. To observe the bands of the amplified DNA products, the products were run in agarose gel at 0.8%, stained with ethidium bromide. The estimation of the molecular weights of the amplified products was carried out by comparison with a 1 Kb molecular marker (Promega, USA). Afterwards, the amplified products were viewed using a Molecular Imager® Gel Doc™ XR+ photodocumenter (BioRad, USA). PCR products were purified using the Wizard® SV Gel kit and the PCR Clean-Up System (Promega, USA) following the manufaturer’s instructions. Sequencing was carried out in the National Genomics Laboratory for Biodiversity, CINVESTAV Irapuato, using oligonucleotides ITS1 (5’-TCCGTAGGTGAACCTGCGG-3’) and ITS4 (5’TCCTCCGCTTATTGATATGC-3’) (White et al.,1990) for the ITS region, and for region D1/D2 28S (LSU), oligonucleotides NL1 (5′-GCATATCAATAAGCGGAGGAAAAG3’) (O’Donnell, 1993) and LR3 (Vilgalys and Hester, 1990); the sequencing process was carried out for both directions of the amplicon.
Phylogenetic analysis of the sequences
The sequences were edited using the program Bioedit Sequence Alignment Editor, version 7.2.5. (Hall, 1999). The alignment was carried out using the program ClustalW, and the consensus sequences obtained were compared with the basic local alignment search tool BLASTN of the NCBI (National Center for Biotechnology Information). The perform the evolutionary analyses, the sequences obtained were saved in the FASTA format, and the alignment was carried out using the program ClustalW included in the software MEGA 6.0 (Tamura et al., 2013). The evolutionary history was inferred using the Maximum Likelihood Method, based on the Tamura-Nei model (Tamura and Nei, 1993). The graphic representation of the dendrogram based on region ITS and 28S (LSU) was carried out with the Neigborh-Joininig method, with a bootstrap analysis of 1000 repetitions to determine the trust values for the clades (Felsenstein, 1985), using reference strains available from the GenBank of the NCBI.
RESULTS
Out of the 4 pieces plated for every diseased fruit, 100% corresponded to the fungus reported for each case. Twenty eight isolates were obtained, related to soft rot in papaya fruits, 4 from Colima, 23 from Veracruz, and 1 from Oaxaca. All isolates were characterized morphologically and deposited in the fungal collection of the CIAD Plant Pathology Laboratory. The isolates from Colima, Oaxaca, and 5 representative from Veracruz were selected for the evaluation of pathogenicity and molecular characterization (Table 1).
Table 1 Species of mucoralean fungi causing soft rot in papaya fruits. Each strain (fungal collection code HP “Papaya Fungus”) corresponds to the fungus obtained from the 4 pieces of tissue plated in PDA per diseased fruit.

x representative isolates used in molecular characterization.
Pathogenicity tests
All Maradol papaya fruits inoculated through lesions with the mucoralean species presented, 24 h after inoculation, symptoms that included humid-looking lesions of approximately 1 cm in diameter; white mycelia developed 48 h later, and after 72 h, sporangia were produced. Finally, 5 d after inoculation, the development of abundant light to dark brown mycelia and sporangia (Figures 1E and 1C), and gray to black (Figures 1D and 1F) was observed. By contrast, the fruits without lesions did not show symptoms of disease (Figures 1B and 1G). Internal damage in the infected fruit consisted of a soft rot that caused detachment of tissues (Figure 1H).
Morphological characteristics of Mucor irregularis
Cultures from isolation HP160, identified as M. irregularis, were circular, uniform, and with abundant, pale yellow aerial mycelia (Figure 2A), a color which was also observed on the reverse side of the plate (Figure 2B). The rate of mycelial growth was 1.48 cm/day at 25 °C; it did not develop at 4 °C, but it did between 10 and 40 °C (Table 2). The sporangiophores, mostly not branched, presented globular sporangia on their tips, of less than 80 μm, and which contained hundreds of elliptical sporangiospores inside them (Figure 2E and 2C). The columella with an almost spherical hyaline appearance presented a collarette, remanent of the sporangial wall (Figure 2D).
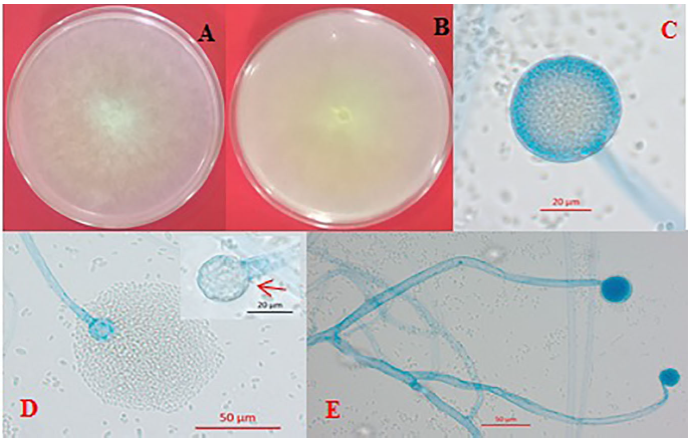
Figure 2 Morphological characteristics of Mucor irregularis. (A) Circular, pale yellow mycelial growth and abundant aerial mycelia, (B) Inverted plate showing a light-yellow color, (C) Globose sporangium with sporangiospores, (D) Release of sporangiospores and presence of columella with an almost spherical shape and a distinctive collarette, and (E) Non-branched hyalin sporangiophores.
Morphological characteristics of Gilbertella persicaria
G. persicaria displayed two types of cultural morphology (Figures 3A and 3E). In the first one, the culture is petaloid with yellow to brown sporangia (Figure 3C), and with a mycelial growth of 1.48 cm/day at 25 °C (strains HP09 and HP114), whereas the second type presents circular cultures, with a more dense mycelia and gray to black sporangia (Figures 3G and 3H), with a mycelial growth rate of 2.46 cm/día (HP15, HP19, HP167, HP178, HP183 and HP211). None of the strains grew at 4 °C, although they did show development between 10 and 40 °C. Other characteristics in G. persicaria include the release of sporangiospores by the breaking of the sporangial wall in two halves (Figure 3I), the presence of small hyalin spines in the walls of the sporangia (Figure 3K), and the presence of hyaline appendages in the sporangiospores (Figure 3J). In the microscopic structures of both types of cultures, no relevant morphological differences were observed (Table 2). The sporangiophores are scarcely branched (four to six in every 100 analyzed); the ovoidal columella with a residual collarette, and cylindrical chlamydospores (Figure 3D), abundant in PDA in 5-day-old cultures.
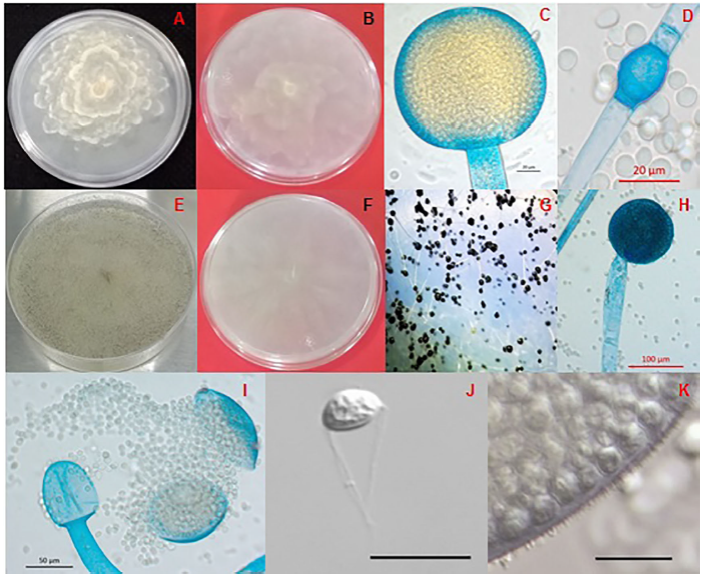
Figure 3 Morphological characteristics of Gilbertella persicaria. (A) Culture in PDA with irregular mycelial growth in petaloid colonies, (B) Reverse side of the culture, (C) Yellowish-brown, multispored globose sporangium, (D) Cylindrical chlamydospore, (E) Culture in PDA with a circular myceliar growth, (F) Reverse side of the culture, (G) Black sporangia, (H) Dark mature multispored globose sporangium, (I) Opening of the sporangial wall in two halves, showing the ovoidal columella, (J) Sporangiospore with polar hyalin appendages, (K) Sporangial wall covered with small hyalin spines. Figures (J and K) bar=20 μm.
Morphological characteristics of Rhizopus oryzae (syn. R. arrhizus)
Cultures of strain HP25 identified as Rhizopus oryzae showed an abundance of initially white aerial mycelia that later turned grayish (Figure 4A and 4B), dark brown to black sporangia that contained sporangiospores with a striated appearance (Figure 4C and 4E). Upon the release of sporangiospores, the light brown globular columella was observed (Figure 4D). The sporangiophores emerge from basal rhizoids (Figure 4F). R. oryzae presented a mycelial growth of 2.46 cm/day at 25 °C; it did not grow at 4°C, but it did between 15 and 40°C (Table 2).
Molecular identification
The reaction of amplification with the oligonucleotides V9G and LR3 generated an approximate fragment of 1500 pb, which included the complete region of ITS1-5.8S-ITS2 and the region D1/D2 of the ribosomal subunit 28S (LSU). The consensus sequences obtained varied for the region ITS1-5.8S-ITS2 from 543 to 751 pb and for the partial region of ribosomal subunit 28S (LSU) from 632 to 711 pb.
The comparison in the data base showed, for the sequencing of the strain HP160 (Access N° KR076763 and KR076764), a percentage of identity of 99% and 96% for the region ITS with strains identified as Mucor sp. (Access N° HM770967 and KP714393, respectively), showing an identity of 91% with the strain of M. irregularis (Access N° JX976251) (Figure 5). However, for the partial ribosomal region 28S (LSU), it showed an identity of 99% with different strains of M. irregularis, which show an alignment of 100% (Figure 6).
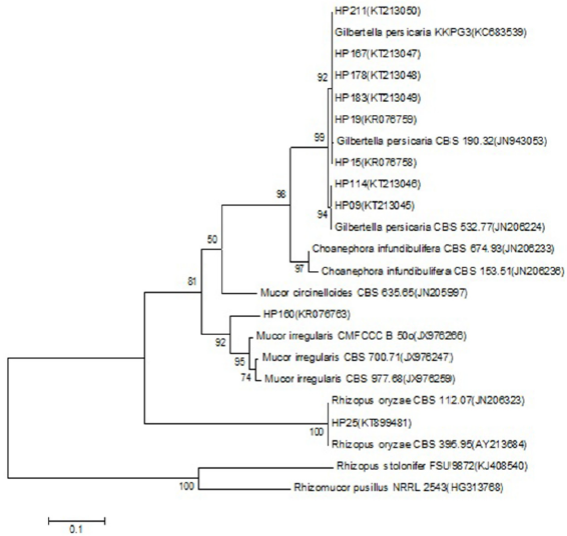
Figure 5 Dendrogram based on the method of highest verosimilitude from sequences obtained for the region ITS1-5.8S-ITS2 of isolations of species of Mucorales and NCBI reference strains; the Access N° are in parentheses. The probability values of the nodes for each clade indicate the percentage of the replications of the analysis of 1000 bootstraps that support that clade. Rhizomucor pusillus was used as an outgroup. The scale represents the number of substitutions per site.
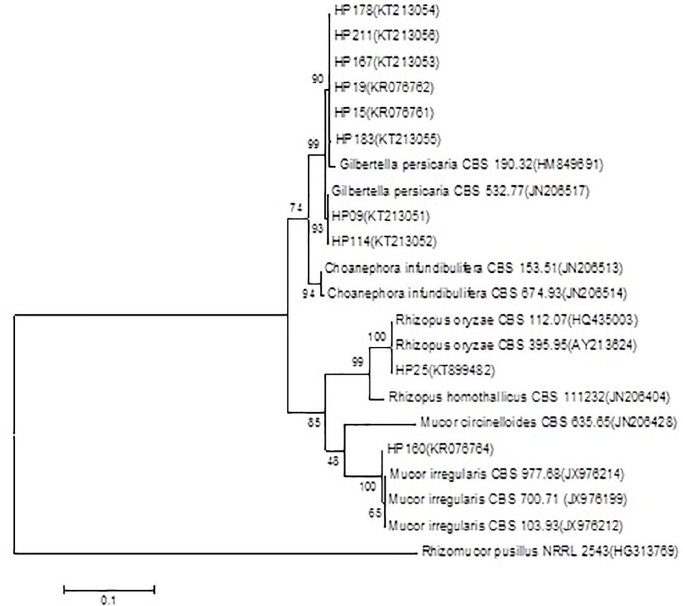
Figure 6 Dendrogram based on the method of highest verosimilitude from the region 28S (LSU) for the strain HP160 and NCBI reference strains; the Access N° are in parentheses. The probability values of the nodes for each clade indicate the percentage of the replications of the analysis of 1000 bootstraps that support that clade. Rhizomucor pusillus was used as an outgroup. The scale represents the number of substitutions per site.
The comparison of the sequences of the region ITS1-5.8S-ITS2 of the strains HP09, HP15, HP19, HP114, HP167, HP178, HP183, and HP211 showed an identity of 100% with sequences deposited in the NCBI (Access N° JN206224 y KC683539) of the species G. persicaria, where we can observe in the phylogenetic analysis the formation of two groups that correspond to the two morphotypes found (Figure 5). The confirmation of this species was carried out by analyzing the sequences of the partial ribosomal region 28S (LSU) (Table 1), which also showed an identity of 100% with the strains (Access N° JN206517 and JN939197) of G. persicaria. Finally, the comparison of the sequence of the strain HP25 of the region ITS1-5.8S-ITS2 and partial region 28S (LSU) (Access N° KT899481 and KT899482) showed an identity of 100% with sequences published in the NCBI (Access N° KJ417552 and AY213624), respectively, of the species R. oryzae (Figure 5). The coverage of the consultations of all the consensus sequences in the NCBI data base varied between 91 and 100%.
DISCUSSION
In Mexico, the identification of plant pathogenic mucoralean species is scarce. The results of this study, focused on using morphological and molecular techniques to identify fungi, showed the presence of three species of mucorales related to the soft rot of papaya fruits. The symptoms produced in papaya fruits were similar to those reported storage polysaccharides (Alves et al., 2002; Krisch et al., 2010).
The morphological characteristics of M. irregularis correspond with descriptions by different authors (Walther et al., 2013; Peng et al., 2015). It is important to mention that, considering that the genus Mucor is polyphyletic, the ITS region was insufficient to discriminate between widely related species, therefore the species was confirmed with the phylogenetic analysis of region 28S (LSU) (Walther et al., 2013; Hoffmann et al., 2013; Hyde et al., 2014). M. irregularis was recently reported as pathogenic in maize (Peng et al., 2015). This is considered a first report of M. irregularis affecting papaya.
The morphology of G. persicaria structures is similar to that reported by diverse authors (Benny, 1991; Guo et al., 2012; Pinho et al., 2014). It is considered a monotypic species in its genus, which is located in the subfamily Gilbertelloideae and the family Choanephoraceae (Hyde et al., 2014). This study observed the development of two morphotypes, which have been defined by Lacap et al. (2003) as strains of the same species that display differences in culture morphologies and rate of mycelial growth, which was confirmed with the separation in clades in the dendrogram of both the ITS and 28S regions (Figures 5 and 6). Morphotype 1 (petaloid colonies, brown sporangia) was observed in 31% of the isolates, while the rest of the isolates presented morphotype 2 (69%). G. persicaria is a common pathogen in tropical and subtropical regions, causing the soft rot of tomato, pear, peach, pitahaya, jambolan (Ginting et al., 1996; Guo et al., 2012; Pinho et al., 2014) and recently, it was reported to have infected papaya fruits in the state of Colima, Mexico (Cruz-Lachica et al., 2016). This study reports the presence of this species in the states of Oaxaca and Veracruz, showing that it is present in other papaya producing regions and its distribution should be considered important.
The morphological characteristics of R. oryzae correspond with those reported by diverse authors, and its identity was confirmed by phylogenetic analysis. R. oryzae has been reported to cause soft rot in banana and citron (Kwon et al., 2012; Hakim et al., 2015), and therefore this work is the first report affecting papaya in Mexico.
According to the origin of the samples analyzed, M. irregularis was found in the state of Veracruz, R. oryzae in the state of Colima, and G. persicaria in Colima, Oaxaca, and Veracruz. In Mexico, Mucor hiemalis and M. circinelloides have been described as affecting papaya in Veracruz (Suárez-Quiroz et al., 2013). However, the identification of species in this study only involved morphological characteristics, therefore performing molecular analyses is recommendable for the following reasons: M. hiemalis and M. irregularis are found in a group of widely related species that require the use of molecular analyses in at least two regions of the genome for their identification (Walther et al., 2013). In the case of M. circinelloides, the identification based on symptoms in fruit and characteristics of sporangia corresponds with results obtained for G. persicaria, therefore distinctive characteristics of the latter species such as the opening of the sporangial wall in two halves and hyalin appendages in sporangiospores can go unnoticed.
CONCLUSIONS
This study showed that Gilbertella persicaria, Mucor irregularis, and Rhizopus oryzae are causal agents of soft rot in papaya fruits. This is the first report on Mucor irregularis and Rhizopus oryzae affecting this crop in Mexico, and it shows that G. persicaria affects fruits in orchards in all three states. Finally, this study shows that the phenotypic and genotypic characterization is, in combination, an adequate strategy to identify species, particularly those that belong to widely related taxons. The access numbers for M. irregularis and R. oryzae in the NCBI genbank were KR076763 and KR076764, and KT899481 and KT899482, respectively.
REFERENCES
Alves MH, Campos-Takaki GM, Figueiredo PAL and Milanez AI. 2002. Screening of Mucor spp. for the production of amylase, lipase, polygalacturonase and protease. Brazilian Journal of Microbiology 33:325-330. http://dx.doi.org/10.1590/S1517-83822002000400009. [ Links ]
Beales P. 2012. Detection of fungal plant pathogens from plants, soil, water and air. Pp: 26-52. In: Lane RC, Bealesand PA and Hughes JDK (eds.). Fungal Plant Pathogens. CAB International. U. K. 324 p. [ Links ]
Benny L. 1991. Gilbertellaceae, a new family of the Mucorales (Zygomycetes). Mycologia 83:150-157. http://dx.doi.org/10.2307/3759930. [ Links ]
Campbell CK, Johnson EM and Warnack DW. 2013. Identification of Pathogenic Fungi. Second Edition. Wiley-Blackwell. Health Protection Agency. U.K. 337 p. [ Links ]
Chukwuka KS, Okonko IO and Adekunle AA. 2010. Microbial ecology of organisms causing pawpaw (Carica papaya L.) fruit decay in Oyo State, Nigeria. American-Eurasian Journal of Toxicological Sciences 2:43-50. http://www.idosi.org/aejts/2(1)10/7.pdf. [ Links ]
Cruz-Lachica I, Marquez-Zequera I, Garcia-Estrada RS, Carrillo-Fasio JA, Leon-Felix J and Allende-Molar R. 2016. First report of Gilbertella persicaria causing papaya fruit rot. Plant Disease 100:227. http://dx.doi.org/10.1094/PDIS-05-15-0607-PDN. [ Links ]
de Hoog GS and van den Ende GAH. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41:183-189. http://dx.doi.org/10.1111/j.1439-0507.1998.tb00321.x. [ Links ]
Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. http://dx.doi.org/10.2307/2408678. [ Links ]
Food and Agriculture Organization (FAO). 2014. Estadísticas. http://faostat3.fao.org/browse/Q/QC/S . Consulta (Julio, 2016). [ Links ]
Ginting C, Zehr EI and Westcott SW. 1996. Inoculum source and characterization of isolates of Gilbertella persicaria from peach fruit in South Carolina. Plant Disease 80:1129-1134. https://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1996Articles/PlantDisease80n10_1129.pdf. [ Links ]
Guo LW, Wu XY, Mao ZC, Ho HH and He YQ. 2012. Storage rot of dragon fruit caused by Gilbertella persicaria. Plant Disease 96(12):1826. http://dx.doi.org/10.1094/PDIS-0712-0635-PDN. [ Links ]
Hakim S, Naz S, Gul S, Chaudhary HJ and Munis MFH. 2015. First report of Rhizopus oryzae causing fruit rot of Citrus medica L. in Pakistan. Journal of Plant Pathology 97:209-220. http://dx.doi.org/10.4454/JPP.V97I1.035. [ Links ]
Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic acids. Symposium Series. Oxford University Press 41:95-98. http://jwbrown.mbio.ncsu.edu/JWB/papers/1999Hall1.pdf. [ Links ]
Hoffmann K, Pawłowska J, Walther G, Wrzosek M, Hoog GS, Benny GL, Kirk PM and Voigt K. 2013. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30:57-76. http://dx.doi.org/10.3767/003158513X666259. [ Links ]
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE et al. 2014. One stop shop: backbones trees for important phytopathogenic genera: I (2014). Fungal Diversity 67:21-125. http://dx.doi.org/10.1007/s13225-014-0298-1. [ Links ]
Krisch J, Takó M, Papp T and Vágvölgyi C. 2010. Characteristics and potential use of β-glucosidases from Zygomycetes. Pp: 891-895. In: Méndez-Vilas A (ed.). Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology. Vol. II. Formatex Research Center. Extremadura, Spain. 1620 p. [ Links ]
Kwon J, Ryu J, Chi TTP, Shen S and Choi O. 2012. Soft rot of Rhizopus oryzae as a postharvest pathogen of banana fruit in Korea. Mycobiology 40:214-216. http://dx.doi.org/10.4489/MYCO.2011.39.2.140. [ Links ]
Lacap DC, Hyde KD and Liew ECY. 2003. An evaluation of the fungal ‘morphotype’ concept based on ribosomal DNA sequences. Fungal Diversity 12: 53-66. http://www.fungaldiversity.org/fdp/sfdp/FD12-53-66.pdf [ Links ]
Michailides TJ. 1991. Characterization and comparative studios of Mucor isolates from stone fruits from California and Chile. Plant Disease 75:373-380. https://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1991Articles/PlantDisease75n04_373.pdf. [ Links ]
Michailides TJ and Spotts RA. 1990. Postharvest diseases of pome and stone fruits caused by Mucor piriformis in the Pacific Northwest and California. Plant Disease 74:537-543. http://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1990Articles/PlantDisease74n08_537.PDF. [ Links ]
Morton J. 1987. Papaya (Carica papaya L.). Pp: 336-346. In: Morton JF (ed.). Fruits of warm climates. Creative Resource Systems Inc. Winterville, USA. 505 p. [ Links ]
O’Donnell K. 1993. Fusarium and its near relatives. Pp: 225-233. In: Reynolds DR and Taylor JW (eds.). The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. CAB International. Wallingford. UK. 375 p. [ Links ]
Papp T, Vastag M, Michailides TJ, Ferenczy L and Vágvölgyi C. 2001. Genetic variability of the postharvest pathogen Gilbertella persicaria: identification of randomly amplified polymorphic DNA (RAPD) markers correlating with (+) and (-) mating types. Antonie Van Leeuwenhoek 80:301-309. http://dx.doi.org/10.1023/A:1013066024258. [ Links ]
Peng XD, Huang SL and Lin SH. 2015. First report of corn kernel brown spot disease caused by Mucor irregularis in China. Plant Disease 99:159-160. http://dx.doi.org/10.1094/PDIS-08-14-0814-PDN [ Links ]
Pinho DB, Pereira OL and Soares DJ. 2014. First report of Gilbertella persicaria as the cause of soft rot of fruit of Syzygium cumini. Australasian Plant Disease Notes 9:143-146. http://dx.doi.org/10.1007/s13314-014-0143-0. [ Links ]
Ryan M, Ritchie BJ and Smith D. 2012. Maintenance and storage of fungal plant pathogens. Pp: 223-250. In: Lane CR, Beales PA and Hughes KJD. Fungal Plant Pathogens. CAB International. South Asia. 324 p. [ Links ]
Suárez-Quiroz ML, Mendoza-Bautista I, Monroy-Rivera JA, de la Cruz-Medina J, Angulo-Guerrero O y González-Ríos O. 2013. Aislamiento, identificación y sensibilidad a antifúngicos de hongos fitopatógenos de papaya cv. Maradol (Carica papaya L.). Revista Iberoamericana de Tecnología Postcosecha 14:115-124. http://www.redalyc.org/pdf/813/81329290004.pdf. [ Links ]
Tamura K and Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10:512-526. http://mbe.oxfordjournals.org/content/10/3/512.long. [ Links ]
Tamura K, Stecher G, Peterson D, Filipski A and Kumar S. 2013. Mega 6: Molecular evolutionary genetics analysis Versión 6.0. Molecular Biology and Evolution 30:2725-2729. http://dx.doi.org/10.1093/molbev/mst197. [ Links ]
Vilgalys R and Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172:4238-4246. http://jb.asm.org/content/172/8/4238. long. [ Links ]
Voigt K, Cigelnik E and O’Donnell K. 1999. Phylogeny and PCR identification of clinically important zygomycetes based on nuclear ribosomal-DNA sequence data. Journal of Clinical Microbiology 37(12):3957-3964. http://jcm.asm.org/content/37/12/3957.long. [ Links ]
Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, Dolatabadi S, Chakrabarti A and de Hoog GS. 2013. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30:11-47. http://dx.doi.org/10.3767/003158513X665070. [ Links ]
White T, Bruns T, Lee S and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetic. Pp: 315-322. In: Innis MA, Gelfand DH, Sninsky JJ and White TJ (Eds.). PCR Protocols: a Guide to Methods and Applications. San Diego. Academic Press. 392 p. https://nature.berkeley.edu/brunslab/papers/white1990.pdf. [ Links ]
Acknowledgements
To the project 2011-163213 “El manejo integral del cultivo de papaya en México, un acercamiento innovador” (The comprehensive management of papaya crops in Mexico, an innovative approach) by SAGARPA for the funding for this research. To CONACyT for the financial support of I. Cruz-Lachica’s studies.
Received: November 08, 2016; Accepted: July 03, 2017











 texto en
texto en