Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.35 n.3 Texcoco Sep. 2017
https://doi.org/10.18781/r.mex.fit.1703-8
Scientific articles
Detection, identification and phylogenetic inference of the stem nematode Ditylenchus dipsaci (Kuhn) filipjev (Nematoda:Anguinidae) affecting alfalfa Medicago sativa L. in Jalisco, Mexico
1Programa de Fitopatología, Colegio de Postgraduados. Km. 36.5 Carretera México-Texcoco, Montecillo, Texcoco, Estado de México. C.P. 56230, México
2CONACYT-Centro Nacional de Metrología, Km 4.5 Carretera a Los Cués, El Marqués, Querétaro, C.P. 76246, México
3Centro Nacional de Referencia Fitosanitaria, Dirección General de Sanidad Vegetal. SENASICA-SAGARPA, Km. 37.5 Carretera Federal México-Pachuca, Tecámac. Estado de México. C.P. 55740. México
4Departamento de Producción Agrícola y Animal, Universidad Autónoma Metropolitana, Calzada del Hueso No. 1100, Col. Villa Quietud, Coyoacán, Ciudad de México, C.P. 0496. México.
Monitoring regulated pests and diseases that affect crops in Mexico is important to determine its presence and impact. With the purpose of supporting the National Phytosanitary Epidemiological Surveillance System activities, alfalfa plants from Atoyac, Jalisco showing stunting, shortened internodes, buds and leaves deformed as well as change leaf coloration, suspicious symptoms produced by foliar nematodes were analyzed. Roots, leaves + buds and stems tissues were analyzed separately. Classical taxonomy, molecular and phylogenetic analysis of two ribosomal DNA markers were performed on specimens detected. Ditylenchus genus specimens were detected and isolated only in leaves + buds and stems. The species identity based on females and males morphotaxonomy belonged to the stem nematode D. dipsaci. The PCR-RFLP restriction pattern of ITS1-5.8S-ITS2 displayed by RsaI enzyme was 335, 280, 200, and 132 bp, while for the enzyme HinfI the pattern was 400 and 300 bp. The BLASTING aligment showed 99% homology with D. dipsaci (E=0.0). The phylogenetic reconstruction of both markers according the bayesian posterior probabilities criteria placed the alfalfa mexican population within the clade that includes several D. dipsaci sensu stricto populations. To our knowledge, this is the first report of D. dipsaci affecting alfalfa in western Mexico.
Key words: Phytosanitary surveillance; taxonomy; phylogeny; molecular; ribosomal DNA
El monitoreo de plagas y enfermedades reglamentadas que afectan a los cultivos en México es importante para determinar su presencia e impacto. Con la finalidad de colaborar con las actividades del Sistema Nacional de Vigilancia Epidemiológica Fitosanitaria, se analizaron plantas de alfalfa de Atoyac, Jalisco con acortamiento de entrenudos, deformación de brotes, hojas y decoloración de foliolos, síntomas sospechosos a infestaciones por nematodos foliares. Se analizaron por separado raíces, hojas + brotes y tallos. Se realizó la taxonomía tradicional, análisis molecular y filogenético de dos marcadores del ADN ribosomal de los nematodos detectados. Se aislaron especímenes del género Ditylenchus solo en hojas + brotes y tallos. La identidad de la especie según la morfotaxonomía de hembras y machos correspondió al nematodo del tallo D. dipsaci. El análisis PCR-RFLP de la región ITS1-5.8S-ITS2 mostró un patrón de restricción de 335, 280, 200 y 132 pb con la enzima RsaI y de 400 y 300 pb con HinfI. La alineación BLAST indicó un 99% de homología con D. dipsaci (E=0.0). La filogenia según el criterio de probabilidades posteriores bayesiano de ambos marcadores ubicó a la población mexicana de alfalfa en el grupo que incluye las poblaciones de D. dipsaci sensu stricto. A nuestro saber, este es el primer reporte de D. dipsaci afectando alfalfa en el occidente de México.
Palabras clave: Vigilancia fitosanitaria; taxonomía; filogenia; molecular; DNA ribosomal
Alfalfa Medicago sativa L. is a forage plant with a high nutritional value in the bovine sector. In Mexico, this crop is affected by biotic factors, among which highlight the pea aphid Acyrthosiphon pisum Harris, the spotted alfalfa aphid Therioaphis maculata Buckton, trips Frankliniella spp., the beet armyworm Spodoptera exigua Hübner, rotting on the base of the stem Rhizoctonia solani Kühn, anthracnosis Colletotrichum trifolii Bain & Essary, Texas root rot Phymatotrichopsis omnivora (Duggar) Hennebert, crown rot (Fusarium spp.), all of which cause production losses (Chew, 2000; Lara and Jurado, 2014). Little is known about the problems caused by nematodes, although in other latitudes they are considered important problems, particularly the root-knot nematodes of the genus Meloidogyne (Griffin and Thyr, 1988; Westerdahl, et al., 2006), lesion nematodes Pratylenchus penetrans (Cobb) Filipjev & Schuurmans Stekhoven (Thies et al., 1992) and the stem nematode Ditylenchus dipsaci (Kühn, 1857) Filipjev 1936 (Wood and Close, 1974; Boelter et al., 1985; Milano de Tomasel and McIntyre, 2001; Perera et al., 2009).
The stem nematode D. dipsaci is considered one of the most important nematological problems in agriculture on temperate climates, standing out crops such as potato (Solanum tuberosum L.), garlic (Allium sativum L.), onion (A. cepa L.), carrot (Daucus carota L.), tulip (Tulipa spp.), clover (Trifolium spp.), beetroot (Beta vulgaris L.), and alfalfa (Medicago sativa L.) (EPPO, 2008; Tenuta et al., 2014). It is a species with obligated endoparasitism, polyphagous of aerial and underground parts of around 500 species of plants, including vegetables, cereals, legumes, and ornamental plants (Caubel and Pedron, 1976; Subbotin and Riley, 2012). It has around 30 physiological races, most of which probably represent species complex that are difficult to differentiate due to their huge morphological, karyoptyic and molecular similarity (Sturhan and Brzesky, 1991; Duncan and Moens, 2006).
The ribosomal DNA analysis has allowed to elucidate the taxonomic situation of D. dipsaci and there is evidence of it being composed by at least seven species: 1. D. dipsaci sensu stricto (s. s.), which is the mos important due to the wide number of crops it affects; 2. D. gigas, known as a “giant” nematode, due to its large size, and it affects Vicia faba; 3. D. weischeri, which affects Cirsium arvense; 4. Ditylenchus sp. D., which affects Pilosella spp.; 5. Ditylenchus sp. E., which affects Crepis praemorsa; 6. Ditylenchus sp. F., which affects Leontodon autumnalis and Pilosella officinarum; and 7. Ditylenchus spp G., which affects Plantago maritima (Subbotin et al., 2005; Kerkoud et al., 2007; Marek et al., 2010; Vovlas et al., 2011; Jeszke et al., 2014).
The physiological race is an intraspecific division that is set apart from other members of the same species by the pathogenicity it causes in different hosts. Races of D. dipsaci s. s. contribute in the wide adaptability of the species to diverse climates and hosts, and is therefore considered of quarantine importance by international plant health regulation agencies (Tenuta et al., 2014).
Alfalfa is a host also affected by D. dipsaci, where it is known as the alfalfa nematode, since sprouts, stems and leaves are infected and can cause defoliation and death of plantlets (Milano de Tomasel and McIntyre, 2001; Westerdahl, et al., 2006). By affecting the botanical and vegetative seed, it is a risk for the dissemination to areas where it is not found (Wood and Close 1974; Gray et al., 1994; Milano de Tomasel and McIntyre, 2001).
In 2014, the National Phytosanitary Epidemiological Surveillance Program (Programa Nacional de Vigilancia Epidemiológica Fitosanitaria - Pro VEF) of the SENASICA-SAGARPA in Jalisco, Mexico, carried out an alfalfa crop sampling by selecting plants with reduced growth, shortening of internodes, distortion and sprouts and leaves discolored. These symptoms are similar to those produced by foliar nematodes in alfalfa (Evans et al., 2008), but in Mexico there was no evidence of its presence, hence the aim of determining the taxonomic and molecular identities of the nematodes found, as well as their phylogenetic affinities.
MATERIALS AND METHODS
Sampling of biological material
Sampling was carried out in the alfalfa crop var. Atoyac with a spraying irrigation system in Cuyacapan, Atoyac, Jalisco between coordinates 19.974489 °N - 103.5276407 °W. Seven samples of complete plants with roots, stems, sprouts, and leaves with symptoms related to foliar nematodes were collected randomly.
Extraction of nematodes
Roots and foliage of the samples were dissected 1-2 cm long, and separated on roots, leaves+sprouts and stems. Samples were incubated separately for 4 h in sterile distilled water at 20±2 °C. The nematodes extracted from the tissues were inspected using a sterescopic American Optical microscope with magnifications 1-15X.
Taxonomic identification
The identity of the species related to the symptom was determined using tradicional taxonomy. Measurements were taken of the diagnosis characteristics, as well as of the calculation of the De Man ratio Indices, and compared with the species reference values for the genus Ditylenchus mentioned by Hooper, 1972; Decker, 1969 and Sturhan & Brzeski 1991. Females and males were anesthesized with the heat from an alcohol burner and then placed on 2% water-agar (Esser, 1986). Photographs were taken using an digital AxioCam ICc1 (Carl Zeiss) digital camera adapted to an Axiostar Plus (Carl Zeiss) compound microscope. The measurements were performed using the program AxioVision Rel 4.8 (Carl Zeiss).
DNA extraction
DNA was extracted using the method proposed by Williams et al., 1992 and Thomas et al., 1997 using the specimens previously analyzed using traditional taxonomy. Nematodes were recovered individually from the mounting medium and placed in 10 μl of lysis buffer solution (10 mM Tris-HCl, pH 8.0, 2.5 mM MgCl2, 50 mM KCl, 0.45% Tween 20; 0.05% gelatin) on a clean slide cover. The specimen was smashed using a micropipette tip under the stereoscopic microscope. The solution was transferred into a 200 μl centrifuge tube and frozen at -40°C for 30 min. It was incubated at 65°C for 1 h, vortexed at least once, and 0.1 μl of proteinase K (20 μg/ml) was added with 10-15 min remaining. Finally, it was incubated at 95 °C for 15 min to inactivate the Proteinase K and stored at -20 °C. For each PCR reaction, 5 μl of the crude DNA extract were used.
Amplification by PCR of ribosomal DNA
Two molecular markers of the ribosomal DNA were used: 1. ITS1-5.8S-ITS2 region, using primers AB28 (5´- GTT TCC GTA GGT GAA CCT GC-3´) (Joyce et al., 1994) and TW81 (5´-ATA TGC TTA AGT TCA GCG GGT-3´) (Howlett et al., 1992). 2. D2-D3 expansion segments of 28S gene, using primers D2A (5`-ACA AGT ACC GTG AGG GAA AGT TG-3´) and D3B (5´-TCG GAA GGA ACC AGC TAC TA-3´) (Ellis et al., 1986; Courtright et al., 2000). For both markers, DNA from D. dipsaci affecting garlic was used as a positive control.
For the amplification of ITS1-5.8S-ITS2, the reaction was composed by PCR 1X (Invitrogen) buffer solution, MgCl2 3 mM, Mix dNTP´s 200 μM, primers TW81 and AB28 at 0.4 μM, Taq Polymerase 2.5 U/μl and 5.0 μl DNA (crude extract). The PCR reactions took place in a total volume of 50 μl in an i-Cycler (BioRad) thermocycler with the following thermocycling program: initial denaturalization at 95 °C for 3 min, 34 denaturalization cycles at 95 °C for 45 s, annealing at 57 °C for 30 s, extension at 72 °C for 1.5 min, and finally, a final extension at 72 °C for 5 min (Skantar et al., 2007).
The amplification components of the expansion segments D2-D3 of gene 28S were: PCR 1X (Invitrogen) buffer solution, MgCl2 3 mM, Mix dNTP´s 200 μM, primers D2A and D3B at 0.4 μM, Taq Polymerase 2.5 U/μl and 5.0 μl DNA, considering a final volume of 50 μl. The thermocycling conditions were: initial denaturalization 95 °C for 3 min, 34 denaturalization cycles 95 °C for 45 s, annealing 55 °C for 45 s, extension at 72 °C for 1 min, and a final extension 72 °C for 5 min (Marek et al., 2010).
The products of PCR from both markers were run by horizontal electrophoresis in 1.4% agarose gel stained with GelRed (Biotium) and visualized in a Gel Doc EZ (BioRad) photodocumenter.
PCR-RFLP’s of the ITS1-5.8S-ITS2 region
The amplified products were digested with restriction enzymes RsaI and HinfI. The final reaction volume was 20 µl with the following components: 2 µl of digestion buffer solution, 1 µl of the enzyme, 10 µl of PCR product and 7 µl of sterile molecular biology grade water.Samples were incubated at 37 °C for 3 h and analyzed by horizontal electrophoresis in 2.5% ultrapure agarose gel-1000 stained with GelRed (Biotium) and visualized in the Gel Doc EZ (BioRad) photodocumenter.
Sequencing, bioinformatics, and phylogenetic analysis
The PCR products amplified were sequenced using the Sanger method at National Phytosanitary Reference Center, SENASICA-SAGARPA. The “forward” and “reverse” sequences were assembled and edited, and primer sequences were eliminated using the program CodonCode Aligner 4.2.7 (CodonCode Co.).
Both ribosomal markers underwent a BLAST to determine homology with NCBI sequences. Phylogenetic analysis was carried out with a multiple alignment of 36 sequences of ITS1- 5.8S-ITS2, whereas D2-D3 expansion segments of the gene 28S, multiple alignment included 28 sequences of species closely with the software CLUSTAL W 2.3 (Thompson et al., 1997). For both phylogenetic reconstructions, D. destructor was used as an external group. The search for the best model of nucleotidic substitution was carried out on Mr Modeltest 2.3 (Nylander, 2004) and PAUP 4 (Swofford, 1998). For the Bayesian inference, the program used was MrBayes v.3.2 (Huelsenbeck and Ronquist 2001) with four chains for 1,000,000 generations. In the Marcov chains, samples were taken at intervals of 100 generations. For each analysis, two runs were performed using the model HKY+G for both markers. The first 20% of the trees sampled were eliminated from all the analyses and the remaining trees were used to calculate the consensus tree by the rule of the majority of 50% and posterior probabilities were calculated for each clade. The trees were visualized using the software FigTree v1.3.1 (Rambaut, 2009).
RESULTS AND DISCUSSION
All plants showed symptoms of growth reduction, shortening of internodes, and deformation of sprouts and leaves (Figure 1A) as well as decoloring of leaflets (albino leaves or “white flagging”) (Figure 1B). These symptoms, and particularly albino leaves, are similar to those caused by the stem nematode D. dipsaci, also been reported in the provinces of Alberta and British Columbia, Canada (Vrain and Lalik, 1983), and the states of Washington, Nevada, Utah, Alabama, and Wyoming, in the United States (Gray et al., 1994).

Figure 1 Symptoms produced by the stem nematode D. dipsaci in alfalfa. A) Deformation of leaves, sprouts and shortening of internodes. B) Decoloring of leaflets (“albino leaves”).
In the incubation of roots, only free living nematodes were isolated, whereas in leaf, sprout and stem tissues, active specimens of the genus Ditylenchus were obtained. Although live nematodes were found (~90-100/g of sample), specimens were also found in anhydrobiosis stage (~15-20). Considering that this species can survive in this way on harvest residues, botanical seeds, and soil particles, the specimens may reactivate and serve as a source of inoculum for future infections or introduce itself into areas in which it was not previously present (Wharton and Barrett, 1985).
Taxonomic identification: The specimens found displayed the following characteristics: Females: thin body (Figure 2A), cephalic area slightly flattened, glandular area with a slight or no overlap, muscular middle bulb with a well-defined valve. Small and weak stylet with well-developed nodules (Figure 2B), post-uterine sac with a length ½ of that between the vulva and the anus (Figure 2D). Males: thin body (Figure 2F), relatively smaller than females; simple spicules, moderate gubernaculum, and leptoderan bursa type that covers approximately 75% tail length (Figure 2E). Both in females and males, a lateral field was observed with four incisures (Fig. 2C) and a sharp conical tail ending in the shape of a pencil tip (Figures 2A, E, and F). These morphological characteristics correspond to the genus Ditylenchus. The morphometry and De Man ratio obtained agree with the reference values reported by Decker, 1969; Hooper, 1972 and Sturhan & Brzeski 1991 for the alfalfa nematode D. dipsaci (Table 1).
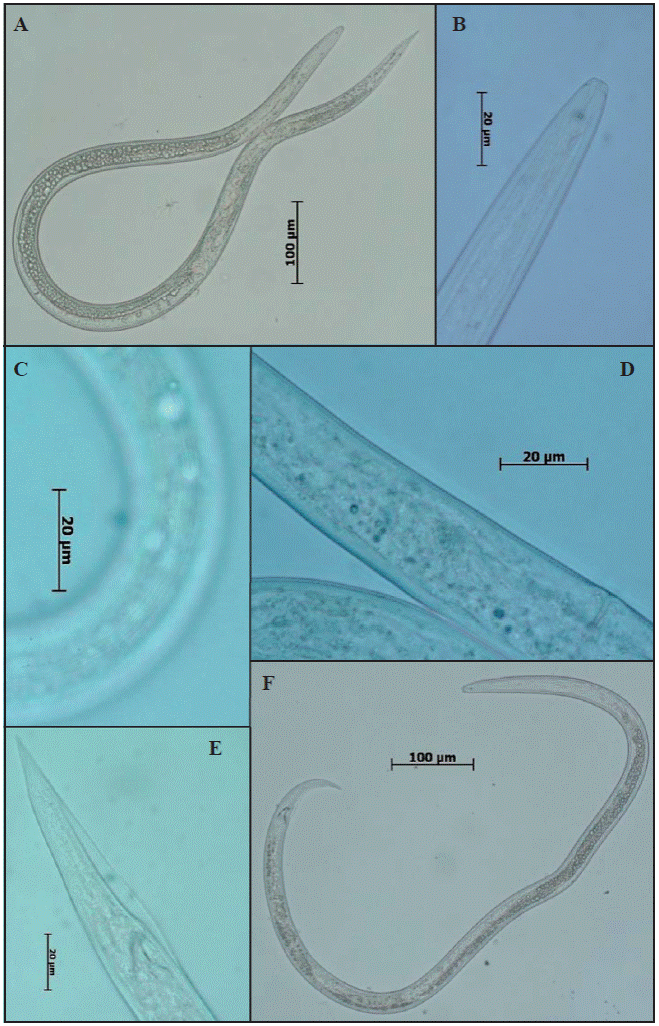
Figure 2 Micrographs of diagnosis characteristics for nematodes found in alfalfa. A) female, B) anterior end and stylet, C) Four lateral fields, D) post-uterine sac, E) sharp conical tail ending and leptoderan bursa type, F) male.
Table 1 Morphometry of Ditylenchus found in alfalfa plants. All values are expressed in mm except V and T, which are expressed as percentages. Average ± Standard deviation and minimum-maximum values.
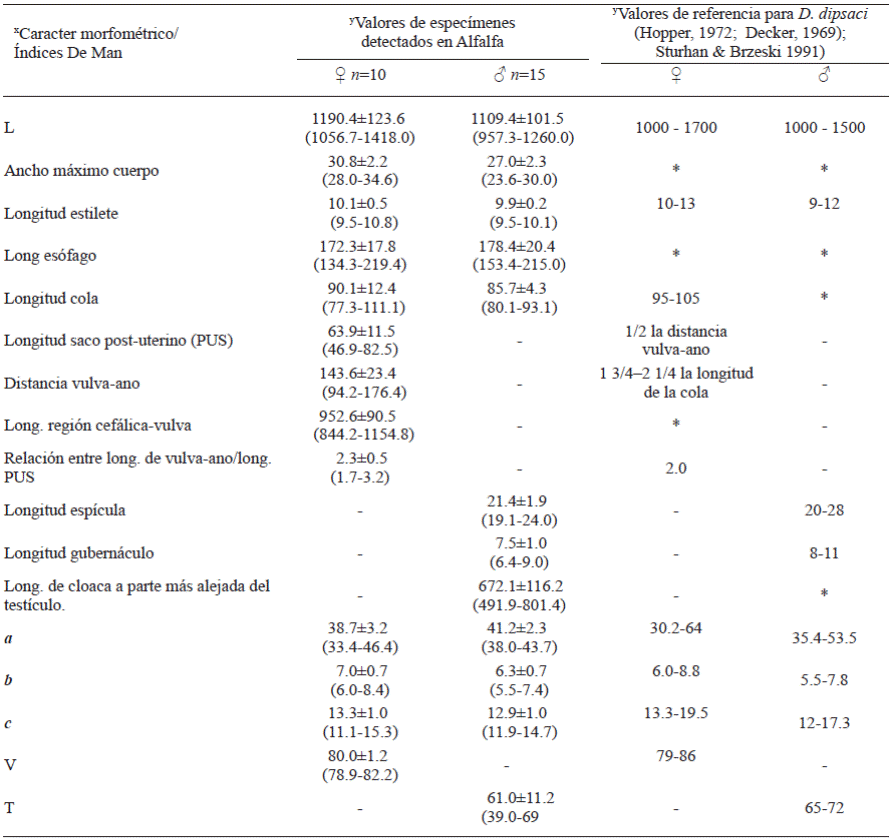
L=Total body length; a=L/Greatest body width; b=L/ Oesophagus length; c=L/tail length; V = Anterior end-vulva length/L x 100; T=Distance from cloaca to anteriormost part of testis/L x100.
♀ = Females, ♂ = Males
y - = Not applicable, * = Value not available in literature.
Molecular identification: The amplification of ITS1-5.8S-ITS2 region and D2-D3 expansion segments of 28S gene of the ribosomal DNA produced a fragment of approximately 750 bp (Figure 3) and 800 bp, respectively (Figure 4), which coincides with that reported by Subbotin et al., 2005. These sequences were deposited in the NCBI database with access numbers KY348762, KY348763, and KY348764 for ITS1-5.8S-ITS2 and KY348765 for D2-D3 of the gene 28S.
PCR-RFLPs enzymatic digestion: The digestion of the PCR product of ITS1-5.8S-ITS2 region with the enzyme RsaI produced four fragments: 335, 280, 200 and 132 bp (Figure 5) and with HinfI, two fragments were produced: 400 and 300 bp (Figure 6). The digestion pattern agreed with the one presented by the bulb nematode D. dipsaci in garlic for both enzymes (Subbotin et al., 2003; Vovlas et al., 2011). However, the digestion pattern for the nematode D. dipsaci found in alfalfa with the enzyme RsaI showed an additional fragment of 200 bp (Figure 5). This 200 bp fragment indicated the differentiation of the population of the nematode found in alfalfa in regard to the population that affects garlic.
Phylogenetic analysis. The search for homology by BLAST of the Mexican sequences of D. dipsaci found in alfalfa with sequences of the NBCI from both markers of the rDNA previously deposited in the GenBank displayed a 99% homology (E=0.0) with D. dipsaci sequences from different hosts and areas.
The topology of the phylogenetic tree obtained with marker ITS1-5.8S-ITS2 positioned D. dipsaci in two large groups, based on karyotypic differences: the diploid and the polyploid. The Mexican population of D. dipsaci in alfalfa was grouped in the clade sensu stricto D. dipsaci diploid (Figure 7), which shows a low haplotype diversity and little intraspecific variation. Although Mexican populations are grouped in a well-defined clade, the presence of different D. dipsaci haplotypes in alfalfa is evident. This clade get together almost all the physiological races of D. dipsaci based on the hosts affected. A similar grouping was observed with marker D2-D3 of the gene 28S (Figure 8). This topology is similar to that reported for members of the Anguinidae family using different rDNA markers (Subbotin et al., 2003; Subbotin et al., 2005; Castillo et al., 2007; Jeszke et al., 2014; Qiao et al., 2016).
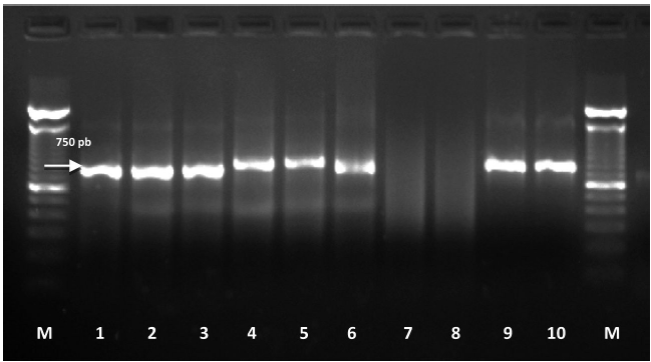
Figure 3 Amplification of rDNA ITS1-5.8S-ITS2 region of D. dipsaci found in alfalfa. M: 100 bp molecular marker; 1-6: amplified from the individual DNA extraction of D. dipsaci; 7 and 8: negative controls (molecular biology grade sterile water); 9 and 10: positive controls for D. dipsaci (garlic) amplifying a fragment of 750 bp.
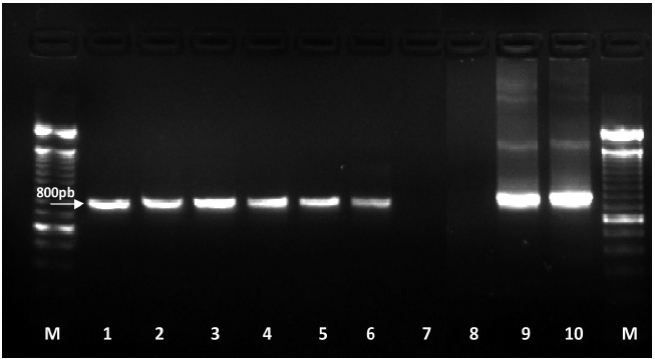
Figure 4 Amplification of D2-D3 expansion segments of 28S gene. M: 100 bp molecular marker; 1-6: amplified from the invidivual DNA extraction of D. dipsaci; 7 and 8: negative controls (molecular biology grade sterile water); 9 and 10: positive controls for D. dipsaci (garlic) amplifying a fragment of 800 bp.
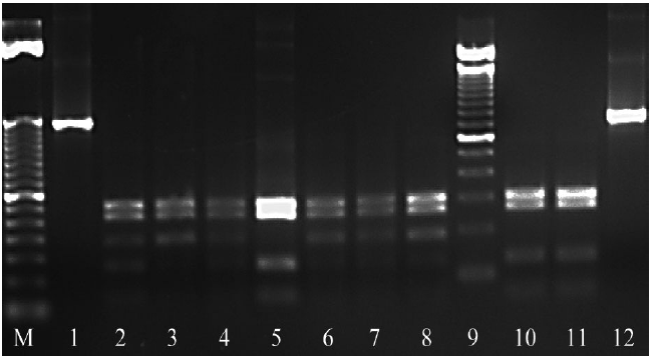
Figure 5 Enzymatic digestion of ITS1-5.8S-ITS2 region of the rDNA with the enzyme RsaI. M: 50 bp molecular marker; 1 and 12: Undigested PCR product; 2-4, 6-8: Restriction pattern 335, 280, 200 and 132 bp of D. dipsaci in alfalfa. 5, 10 and 11: Positive control of D. dipsaci (garlic) restriction pattern 335, 280 and 132 bp: 9: 100 bp molecular marker.
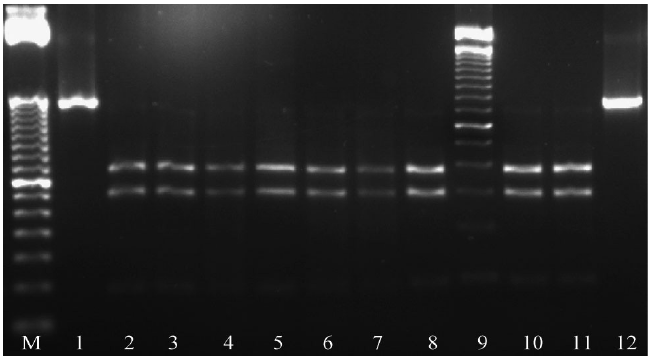
Figure 6 Enzymatic digestion of ITS1-5.8S-ITS2 region of rDNA with the enzyme HinfI. M: 50 bp molecular marker; 1 and 12: Undigested PCR product; 2-8: Restriction pattern 400 and 300 bp, 9: 100 bp molecular marker. 10 and 11: Restriction pattern for D. dipsaci s.s. (garlic) of 400 and 300 bp.

Figure 7 Bayesian phylogenetic reconstruction of ITS1-5.8S-ITS2 region sequences from Mexican population of D. dipsaci found in alfalfa. Superior probability consensus tree 50% generated from the complex model: HKY + G, with other anguinid sequences deposited in GenBank. Support values are indicated in each one of the clades.
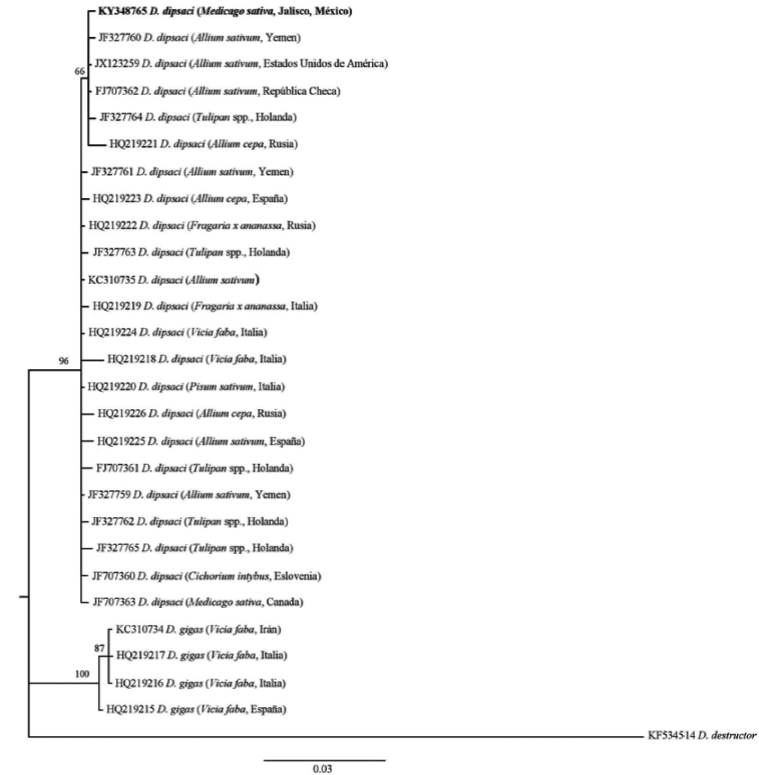
Figure 8 Bayesian phylogenetic reconstruction of D2-D3 expansion segments of 28S gene sequences from Mexican population of D. dipsaci found in alfalfa. Superior probability consensus tree 50% generated from the complex model: HKY + G, with other anguinid sequences deposited in GenBank. Support values are indicated in each one of the clades.
CONCLUSIONS
Mexico, D. dipsaci affects garlic and onion plants in the main geographic areas that produce these alliacous crops (Tenente, 1996; SENASICA, 2013). Studies carried out on differential hosts for the determination of races, claim that such correspond to the “garlic race” (Aguilera, 1994). The pathogenic variability of D. dipsaci manifested in several races is well-known based on the host it affects, and therefore, based on the host and the PCR-RFLP restriction pattern obtained with the endonuclease RsaI, it is posible to indicate that the foliar nematode D. dipsaci found in this host could be the “alfalfa race,” since this race only affects, and reproduces in Medicago sativa plants (Webster, 1967; Sturhan and Brzesky, 1991; Riggs, 1991). It is necessary to carry out studies of differential hosts in the populations that affect alfalfa and use genomic tools as high throughput sequencing (Next Generation Sequencing) to obtain useful molecular markers for the differentiation of the D. dipsaci races in Mexico.
REFERENCES
Aguilera AWH. 1994. Identificación de razas del nematodo Ditylenchus dipsaci presentes en México. Tesis Profesional, UAM-X. 21 p. [ Links ]
Boelter RH, Gray FA and Delaney RH. 1985. Effect of Ditylenchus dipsaci on Alfalfa mortality, winterkill, and yield. Journal of Nematology 17:140-144. http://journals.fcla.edu/jon/article/viewFile/65631/63299 [ Links ]
Castillo P., Vovlas N., Azpilicueta A., Landa B.B. and Jiménez-Díaz R.M. 2007. Host-parasite relationships in fall-sown sugar beets infected by the stem and bulb nematode, Ditylenchus dipsaci. Plant Disease 91:71-79. http://apsjournals.apsnet.org/doi/pdf/10.1094/PD-91-0071 [ Links ]
Caubel G. and Pedron D. 1976. Distribution géographique du nématode des tiges Ditylenchus dipsaci (Kuhn) Fil., en culture de légumineuses fourragéres. Sciences Agronomiques Rennes 8:183-188. [ Links ]
Courtright EM, Wall DH, Virginia RA, Frisse LM, Vida JT and Thomas WK. 2000. Nuclear and mitochondrial DNA sequence diversity in the antarctic nematode Scottnema lindsayae. Journal of Nematology 32: 143-153. http://journals.fcla.edu/jon/article/view/67138/64806 [ Links ]
Chew MYI. 2000. Enfermedades de la Alfalfa. Pp: 47-57. In: Núñez HG, Chew MYI, Reyes JI y Godina GHJ (eds.). Producción y utilización de la alfalfa en la zona norte de México. INIFAP Centro de Investigación Regional Norte Centro. México. 102 p. http://biblioteca.inifap.gob.mx:8080/jspui/bitstream/handle/123456789/1938/produccionyutilizaciondelaalfalfaenlazonanortedemexico.pdf?sequence=1 [ Links ]
Duncan WL and Moens M. 2006. Migratory endoparasitic nematodes. Pp: Perry RN and Moens M. (eds.). 144-166. In: Plant Nematology. CABI, UK. 541p. [ Links ]
Decker H. 1969. Phytonematologie. VEB Deutscher Landwirtschaftsverlag, Berlin, Germany. 526p. [ Links ]
Esser RP. 1986. A water agar en face technique. Proceedings of the Helminthological Society of Washington 53:254-255. Disponible en línea: http://bionames.org/bionames-archive/issn/0018-0130/53/254.pdf [ Links ]
Ellis RE, Sulston JE and Coulson AR. 1986. The rDNA of C. elegans: Sequence and structure. Nucleic Acids Research 14:2345-2364. Disponible en línea: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC339662/pdf/nar00274-0410.pdf [ Links ]
Evans K., Israelsen C. and Pace M. 2008. Alfalfa Stem Nematode. Utah State University Extension and Utah Plant Pest Diagnostic Laboratory. Utah, USA. 3 p. Disponible en línea: https://utahpests.usu.edu/uppdl/files-ou/factsheet/alfalfa-stem-nematode08.pdf [ Links ]
Gray FA, Williams JL, Griffin GD and Wilson TE. 1994. Distribution in the Western United States on alfalfa and cultivar Reaction to mixed populations of Ditylenchus dipsaci and Aphelenchoides ritzemabosi. Journal of Nematology 26:705-719. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619569/pdf/705.pdf [ Links ]
Griffin GD and Thyr BD. 1988. Interaction of Meloidogyne hapla and Fusarium oxysporum f. sp. medicaginis on alfalfa. Phytopathology 78:421-425. https://www.apsnet.org/publications/phytopathology/backissues/Documents/1988Articles/Phyto78n04_421.PDF [ Links ]
Hooper DJ. 1972. Ditylenchus dipsaci. CIH. Description of Plant Parasitic Nematodes. Set 1, No. 14. [ Links ]
Howlett BJ, Brownlee AG, Guest DI, Adcock GJ, and McFadden GI. 1992. The 5S ribosomal RNA gene is linked to large and small subunit ribosomal RNA genes in the oomycetes, Phytopthora vignae, P. cinnamomi, P. megasperma f. sp. glycinae and Saprolegnia ferax. Current Genetics 22: 455-461. DOI: 10.1007/BF00326410 [ Links ]
Huelsenbeck J and Ronquist F. 2001. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755. DOI: https://doi.org/10.1093/bioinformatics/17.8.754 [ Links ]
Jeszke A, Budziszewska M, Dobosz R, Stachowiak A, Protasewicz D, Wieczorek P and Obrępalska-Stęplowska A. 2014. A comparative and phylogenetic study of the Ditylenchus dipsaci, Ditylenchus destructor and Ditylenchus gigas populations occurring in Poland. Journal of Phytopathology 162: 61-67. DOI: 10.1111/jph.12161 [ Links ]
Joyce SA, Reid A, Driver F and Curran J. 1994. Application of polymerase chain reaction (PCR) methods to identification of entomopathogenic nematodes. Pp:178-187. In: Burnell AM, Ehlers RU. and Masson JP. (eds.). COST 812 Biotechnology Genetics of Entomopathogenic Nematode- Bacterium Complexes. Proceedings of Symposium & Workshop, St. Patrick’s College, Maynooth, County Kildare, Ireland. [ Links ]
Kerkoud M, Esquibet M, Plantard O. et al., 2007. Identification of Ditylenchus species associated with Fabaceae seeds based on a specific polymerase chain reaction of ribosomal DNA-ITS regions. European Journal of Plant Pathology 118: 323-332. DOI: 10.1007/s10658-006-9092-6 [ Links ]
Lara MCR, y Jurado GP. 2014. Paquete tecnológico para producir alfalfa en el estado de Chihuahua. Centro de Investigación Regional Norte Centro, INIFAP. Chihuahua, México. 48 p. Disponible en línea: http://siproduce.sifupro.org.mx/seguimiento/archivero/8/2013/anuales/anu_2413-25- 2014-05-6.pdf [ Links ]
Marek M, Zouhar M, Douda O, Mazakova J. and Rysanek P. 2010. Bioinformatics -assisted characterization of the ITS1-5.8S-ITS2 segments of nuclear rRNA gene clusters, and its exploitation in molecular diagnostics of European crop-parasitic nematodes of the genus Ditylenchus. Plant Pathology 59: 931-934. DOI: 10.1111/j.1365- 3059.2010.02322.x [ Links ]
Milano de Tomasel MC, and McIntyre GA. 2001. Distribution and biology of Ditylenchus dipsaci and Aphelenchoides ritzemabosi in alfalfa grown in Colorado. Nematropica 31:11-16. Disponible en línea: http://journals.fcla.edu/nematropica/article/view/69608/67268 [ Links ]
Nylander J. 2004. MrModeltest 2.3. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [ Links ]
Perera M, Taylor S, Vanstone V and Jones M. 2009. Protein biomarkers to distinguish oat and lucerne races of the stem nematode, Ditylenchus dipsaci, with quarantine significance for Western Australia. Nematology 11:555-563. https://doi.org/10.1163/138855409X12465362560557 [ Links ]
Qiao Y, Yu Q, Badiss A, Zaidi MA, Ponomareva E, Hu Y, and Ye W. 2016. Paraphyletic genus Ditylenchus Filipjev (Nematoda, Tylenchida), corresponding to the D. triformis-group and the D. dipsaci-group scheme. ZooKeys 568: 1-12. DOI: 10.3897/zookeys.568.5965 [ Links ]
Rambaut A. 2009. FigTree. v1.3.1. Available: http://tree.bio.ed.ac.uk/software/. [ Links ]
Riggs RD. 1991. Resistance-breaking races of plant parasitic nematodes. Pp. 827-854. In: Manual of Agricultural Nematology. Nickle, W. R. (Ed.). Marcel Dekker Inc. New York, USA. 1025 p. [ Links ]
SENASICA, 2013. Nematodo de los tallos y de los bulbos (Ditylenchus dipsaci Kühn 1857). Dirección General de Sanidad Vegetal-Sistema Nacional de Vigilancia Epidemiológica Fitosanitaria. México. Ficha Técnica No. 18. 24 p. Disponible en línea: http://www.cesaveson.com/files/docs/campanas/FTNo.18nematododeltalloydelosbulbos2.pdf [ Links ]
Skantar AM, Handoo ZA, Carta LK, and Chitwood DJ. 2007. Morphological and molecular identification of Globodera pallida associated with potato in Idaho. Journal of Nematology 39:133-144. Disponible en línea: https://www.ars.usda.gov/ARSUserFiles/990/Skantar%20Chitwood%20Idaho%20Globodera%20pallida%202007reduced.pdf [ Links ]
Sturhan D and Brzeski MW. 1991. Stem and bulb nematodes, Ditylenchus spp. Pp. 423-464. In: Nickle WR (ed.). Manual of Agricultural Nematology. Marcel Dekker, Inc., New York (USA). 1064 p. [ Links ]
Swofford D. 1988. PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version 4.0b8a. Sunderland, MA: Sinauer. [ Links ]
Subbotin SA, Madani M, Krall EL, Sturhan D and Moens M. 2003. Identification and phylogenetic relationships within the stem nematode Ditylenchus dipsaci complex (Tylenchida: Anguinidae) as inferred from analysis of the ITS-rDNA sequences. Journal of Nematology 35:365. Disponible en línea: http://journals.fcla.edu/jon/article/ view/67424/65092 [ Links ]
Subbotin SA, Madani M, Krall E, Sturhan D and Moens M. 2005. Molecular diagnosis, taxonomy, and phylogeny of the stem nematode Ditylenchus dipsaci species complex based on the sequences of the internal transcribed spacer-rDNA. Phytopathology 95:1308-1315. Disponible en línea: http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO-95-1308 [ Links ]
Subbotin SA and Riley IT. 2012. Stem and gall forming nematodes. Pp. 521-577. In: Manzanilla-Lopez RH and Marban-Mendoza N. (eds.). Practical Plant Nematology. Biblioteca Básica de Agricultura. Colegio de Postgraduados (México). 883 p. [ Links ]
Tenente RCV.1996. Nematode problems of bulbs, with special reference to Ditylenchus dipsaci. Nematropica 26:91-99. http://journals.fcla.edu/nematropica/article/ view/64150/61818 [ Links ]
Tenuta M, Madani M, Briar S, Molina O, Gulden R, and Subbotin SA. 2014. Occurrence of Ditylenchus weischeri and not D. dipsaci in field pea harvest samples and Cirsium arvense in the Canadian Prairies. Journal of Nematology 46:376-384. https://www.ncbi.nlm.nih.gov/pmc/articles/ PMC4284090/pdf/376.pdf [ Links ]
Thies JA, Barnes DK, Rabas DL, Sheaffer CC and Wilcoxson RD. 1992. Seeding Date, Carbofuran, and Resistance to Root-Lesion Nematode Affect Alfalfa Stand Establishment. Crop Sciences 32:786-792. DOI:10.2135/cropsci1992.0011183X003200030042x [ Links ]
Thomas WK, Vida JT, Frisse LM, Mundo M and Baldwin JG. 1997. DNA sequences from formalin-fixed nematodes: Integrating molecular and morphological approaches to taxonomy. Journal of Nematology 29:250-254. Disponible en línea: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2619797/pdf/250.pdf [ Links ]
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24:4876-82. DOI: 10.1093/nar/25.24.4876 [ Links ]
Vovlas N, Troccoli A, Palomares-Rius JE, De Luca F, Liébanas G, Landa BB, Subbotin SA and Castillo P. 2011. Ditylenchus gigas n. sp. parasitizing broad bean: a new stem nematode singled out from the Ditylenchus dipsaci species complex using a polyphasic approach with molecular phylogeny. Plant Pathology 60: 762-775. Disponible en línea: http://onlinelibrary.wiley.com/doi/10.1111/j.1365-3059.2011.02430.x/epdf [ Links ]
Vrain TC and Lalik B. 1983. Distribution and pathogenicity of the Alfalfa Stem Nematode, Ditylenchus dipsaci, in British Columbia. Plant Disease 67:300-302. http://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1983Articles/PlantDisease67n03_300.PDF [ Links ]
Webster JM. 1967. The significance of biological races of Ditylenchus dipsaci and their hybrids. Annals of Applied Biology 59: 77-83. DOI: 10.1111/j.1744-7348.1967.tb04418.x [ Links ]
Wharton DA and Barrett J. 1985. Ultrastructural changes during recovery from from anabiosis in the plant parasitic nematode, Ditylenchus. Tissue Cell 17:79-96. http://dx.doi.org/10.1016/0040-8166(85)90017-5 [ Links ]
Wharton DA, Rolfe RN and Perry R.N. 2000. Electrophysiological activity during recovery from anhydrobiosis in fourth stage juveniles of Ditylenchus dipsaci. Nematology 2:881-886. DOI: 10.1163/156854100750112824 [ Links ]
Wendt KR, Vrain TC and Webester JM. 1993. Separation identification of three species of Ditylenchus and some host races of D. dipsaci by restriction fragment length polymorphism. Journal of Nematology 25: 555-563. Disponible en línea: http://journals.fcla.edu/jon/article/view-File/66542/64210 [ Links ]
Westerdahl B, Goodell P, and Hafez S. 2006. Alfalfa nematodes. UC IPM Pest Management Guidelines for alfalfa. Disponible en línea: http://ipm.ucdavis.edu/PMG/r1200111.html [ Links ]
Williams BD, Schrank B, Huynh C, Shownkeen R and Waterston RH. 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131: 609-624. Disponible en línea: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1205034/pdf/ge1313609.pdf [ Links ]
Wood FH and Close RC. 1974. Dissemination of lucerne stem nematode in New Zealand. New Zealand Journal of Experimental Agriculture 2:79-82. DOI:10.1080/03015521.1974.10427674 [ Links ]
Acknowledgements
Received: March 23, 2017; Accepted: June 13, 2017











 text in
text in 


