Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Revista mexicana de fitopatología
On-line version ISSN 2007-8080Print version ISSN 0185-3309
Rev. mex. fitopatol vol.35 n.1 Texcoco Jan. 2017
https://doi.org/10.18781/r.mex.fit.1606-1
Scientific articles
Phytophthora hydropathica and Phytophthora drechsleri isolated from irrigation channels in the Culiacan Valley
1Centro de Investigación en Alimentación y Desarrollo. CIAD AC. Área de Horticultura. Km 5.5 Carretera Culiacán-Eldorado, Campo El Diez. Culiacán, Sinaloa, México. CP 80110 Teléfono 6677605536. (brando.alvarez@estudiantes.ciad.mx, rsgarcia@ciad.mx, jvaldez@ciad.mx, ljosefina@ciad.mx, rallende@ciad.mx)
2Universidad Michoacana de San Nicolás de Hidalgo, Laboratorio de Patología Vegetal, Instituto de Investigaciones Agropecuarias y Forestales. Km. 9.5 Carretera Morelia-Zinapécuaro, Tarímbaro, Michoacán. CP 58880. Teléfono (443) 2958323. (fpavia@umich.mx)
Up to date, there are no reports of the presence of Phytophthora species in surface water bodies in Mexico, which represents a risk for the local agriculture. During January 2015, 25 irrigation channels from Culiacan Valley were sampled for the isolation of Phytophthora spp. Isolates were obtained with rhododendron leaves and pear fruits baits. Twenty-nine isolates of Phytophthora were obtained and identified, by morphological characteristics together with rDNA sequences of internal transcribed spacers (ITS) and elongation factor 1-α (TEF-1α). Pathogenicity tests were performed on tomato and pepper leaves, since they are important crops in the region and they are irrigated with water from the sampled channels. Two Phytophthora species were identified: P. hydropathica and P. drechsleri. Isolates of P. hydropathica caused necrotic lesions on tomato and pepper leaves while isolates of P. drechsleri caused necrotic lesions only on tomato leaves. To our knowledge, this is the first report of P. hydropathica in Mexico.
Key words: irrigation water; oomycetes; bait; detection; pathogenicity
En México, no existen reportes de la presencia de especies de Phytophthora en aguas superficiales, de ahí la importancia de determinarlas ya que representan un riesgo para la agricultura del país. En enero de 2015, se muestrearon 25 canales de irrigación en el Valle de Culiacán con el objetivo de identificar las especies de Phytophthora presentes. Los aislamientos fueron obtenidos con trampas que consistieron en hojas de azalea y frutos de pera. Se obtuvieron 29 aislamientos de Phytophthora, los cuales fueron identificados con base en características morfológicas y en secuencias de ADNr de los espaciadores internos transcritos (ITS) y del factor de elongación 1-α (TEF-1α). Se realizaron pruebas de patogenicidad en hojas de tomate y chile, debido a que son cultivos importantes en la región y son irrigados con agua de los canales muestreados. Se identificaron 2 especies: P. hydropathica y P. drechsleri. Los aislamientos de P. hydropathica ocasionaron lesiones necróticas en hojas de tomate y de chile; mientras que los aislamientos de P. drechsleri solamente causaron lesiones en hojas de tomate. De acuerdo con nuestro conocimiento, este es el primer reporte de P. hydropathica en México.
Palabras clave: agua de riego; oomicetes; cebo; detección; patogenicidad
Phytophthora species have the potential of causing devastating diseases in forest and agricultural ecosystems (Erwin and Ribeiro, 1996), since they are able to infect a wide range of hosts. Such is the case of P. cinnamomi, which can damage approximately 2000 plant species (Hardham, 2005). Another example is Phytophthora sojae, which affects a wide range of hosts and causes economic losses of up to 2 billion dollars worldwide (Tyler, 2007).
The presence of plant pathogenic oomycetes in irrigation water has been widely documented (Hong and Moorman, 2005); specifically, there are around 30 species of the genus Phytophthora,which have been isolated from surface water bodies such as rivers, irrigation channels, recycled waters, and reservoirs (Hüberli et al., 2013; Zappia et al., 2014).
Phytophthora species require aquatic environments in order to develop their asexual life cycle and therefore spread quickly with the formation of sporangia and zoospores; for this reason, they have been regularly isolated from irrigation water channels, water reservoirs, and rivers (Sutton et al., 2009; Ghimire et al.,2009; Hulvey et al., 2010; Bienapfl and Balci et al., 2014). Examples of species reported with this dispersal mean include P. cactorum, P. parasitica, P. citricola, P. gonapodyides, and P. cambivora, isolated from irrigation waters in the state of Washington (Yamak et al., 2002). Recently, one of the species with the greatest incidence in surface waters has been P. hydropathica (Brazee et al., 2016; Ghimire et al., 2011; Hulvey et al., 2010). In most of these cases, the presence of Phytophthora species in water has not been related with the existence of symptoms or damages in terrestrial plant hosts (Resser et al., 2011). In Mexico, the presence of 17 species of Phytophthora have been documented (Fernandez-Pavía et al., 2015), the most studied of which include P. capsici, P. cinnamomi, and P. infestans (Fernandez-Pavía et al., 2013). These species have been identified from isolations obtained from diseased plants, mainly vegetables.
Historically, Phytophthora species have been identified by their morphological characteristics; however, identification is difficult, even for experts, due to the similarities in characteristics between some species. Currently, molecular analyses based on DNA studies have been used to characterize populations of Phytophthora in agricultural fields and in natural ecosystems (Blair et al., 2008; Grünwald et al., 2011). The regions of the genome most used for these studies are the internal transcribed spacer (ITS) (Blair et al., 2008) and some nuclear genes, such as the gene that codifies for the factor of elongation 1-α (TEF-1α) and the β-tubulin gene (Kroon et al., 2004).
The objectives of this study were to identify Phytophthora species from irrigation channels in the Culiacán Valley and to evaluate their pathogenicity in agriculturally important plant in the region.
Materials and methods
Isolate collection
The Phytophthora spp. isolates were collected from 25 irrigation channels in District 010 of the CuliacánValley in January 2015 (Table 1). The sampling sites were chosen at random; the traps used were organza bags closed with nylon thread in the upper section, which also helped tie the traps to a particular point by the side of the channel (Figure 1). Bags contained 1 pear fruit and 3 azalea leaves, neither one showing any signs of diseases or damages, and 2 bags were placed in each sampling site. Plant material was washed using tap water and disinfected with ethanol at 70 % for 30 s before being placed in the traps. The traps remained in the surface of the irrigation channels for 48 h (Bush et al., 2003). Afterwards, the traps were gathered and taken to the laboratory. The isolation was carried out by placing several sections of infected plant tissue (Figure 2) in a selective medium containing V8 juice (200 mL/L), natamycin 10 µg/L, ampicillin 200 µg/L, rifampicin 10 µg/L, pentachloronitrobenzene 25 µg/L, and hymexazol 50 µg/L (V8-PARPH) (Jeffers, 1986). The Petri dishes were incubated at room temperature (25±2 °C) for 3 to 5 days. The Hyphal tips from developing colonies were transferred to obtain pure cultures (Hajebrahimi and Banihashemi, 2011). Finally, the isolations were preserved placing 5-7 mycelium discs in active growth(5 mm diameter) in corn meal agar (CMA) inmicrofuge tubeswith sterile distilled water, which were then stored at 15 °C (Martin et al., 2012).
Table 1. Sampling sites in irrigation channels in the Culiacán Valley, species identified and accessions of the sequences deposites inNCBI.
ND= Not determined
* The accession number corresponds to the sequences obtained with the primers ELONGF1/ELONGR1.
Morphological characterization
Sporangia were produced by taking sections of approximately 10 mm2 of cultures with 3 to 5-day growth in V8 medium; they were then placed in 90 mm Petri dishes and flooded with sterilized rainwater. The dishes were kept at room temperature (25±2 °C) for 24-48 h (Gallegly and Hong, 2008; Bienapfl and Balci, 2014). The sporangia were observed under a Carl Zeiss Axiostar Imager A2 light microscope with an integrated camera and measured using the software ZEN 2012 Blue edition (Carl Zeiss).
Structures such as hyphal swellings, antheridia, oogonia and oospores were observed in clarified V8 agar medium(V8 juice under 4000 rpm for 20 min) in cultures of approximately one month of age, incubated in the dark at 20 °C (Mitchell et al., 1992; Bienapfl and Balci, 2014). To determine the maximum growth temperature, discs of 5 mm in diameter with active growth were placed in V8 medium and incubated at 30, 35, and 40 °C ± 1 °C. Mating type. The isolates were confronted with P. nicotianae reference strains with compatibility types A1 and A2. A section of 5 mm of isolates A1 and A2 of P. nicotianae were transferred to a Petri dish with clarified V8 agar and a disc, 5 mm in diameter, of each of the isolates was placed on the opposite side of the dish. The cultures were incubated at 22 °C for approximately 3 weeks (Bienapfl and Balci, 2014). The morphological characteristics and the dimensions of the antheridia, oogonia, and oospores were recorded for the description of the species. The asexual and sexual structures were compared with a tabular key of Phytophthora species (Martin et al., 2012).
Molecular characterization
Approximately 3 mg of mycelia were used, which grew when placing V8 agar discs with active growth, 10 mm in diameter, of each of the isolates in a potato broth medium, and later left for shaking at 150 rpm. From the mycelium produced, the DNA extraction was carried out. For this, we used a digestion buffer (Tris/HCl, 10 mM, pH 8.0; EDTA, 50 mM; SDS, 0.5 %; Triton X-100, 0.5 %; Tween 20, 0.5 %) and 2 µL of K proteinase (20 mg/mL), followed by a series of washings with chloroformisoamylalcohol (Zelaya-Molina et al., 2011). For the characterization of the isolates, the primers DC6 (5’-GAGGGACTTTTGGGTAATCA-3’) (Bonants et al., 1997), and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) (White et al., 1990) were used to amplify the region beside the ITS of an approximate size of 1300 p band primers ELONGF1 (5´-TCACGATCGACATTGCCCTG-3´) and ELONGR1 (5´-ACGGCTCGAG ATGACCAT G-3´) were used to amplify a fragment of approximately 970 pbof the central region of the gene that codifies for elongation factor 1-α in which introns are not included (Kroon et al., 2004).The final concentration of the mixture for PCR consisted of 1X buffer, dNTPs 0.2 mM, each primer at 0.2 mM, 1 unit of polymerase DNA, for a final volume of 25 µL. Amplification conditions for primers DC6/ITS4 consisted of 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min, and a final extension of 72 °C for 10 min, and for primers ELONF/ELONR, they consisted of 95 °C for 2 min followed by 35 cycles of 95 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min, and a final extension of 72 °C for 10 min. The PCR products were observed in a 1 % agarose gel dyed with red gel (Biotium, USA) under 70 V for 90 min. Later, the PCR products were purified using the kit wizard SV Gel and PCR Clean-Up System (Promega, USA). Both chains of the amplicon were sequenced in LANGEBIO, CINVESTAV, Irapuato. The sequences obtained were edited to obtain consensus sequences in the program Bioedit (version 7.2). The sequences of the isolations were compared with sequences of type isolates deposited in the Phytophthora specialized database (www.phytophthoradb.org) or in the NCBI to determine the percentage of similarity.
Evaluation of pathogenicity
The pathogenicity tests were carried out on Capsicum annum chili pepper leaves (hybrid 5810 Syngenta) and Solanum lycopersicum tomato (var. Horus). The completely developed chili leaves and tomato leaf lets were washed with tap water, dried, and disinfected with ethanol at 70 % for 30 s. Afterwards, four leaves of each material, in which incisions were previously made with a sterile dissection needle, were inoculated artificially in the abaxial section, placing a disc (5 mm diameter) with active growth from both species of Phytophthora, seven days old in PDA medium. One leaf from each plant evaluated was used as a control, in which incisions were also made, and placed in a PDA culture medium disc. All leaves were placed in a wet chamber atroom temperature (25±2 °C) for five days, during which the length of the necrotized area of the leaves was recorded on a daily basis (Orlikowski et al., 2012).
Results and Discussion
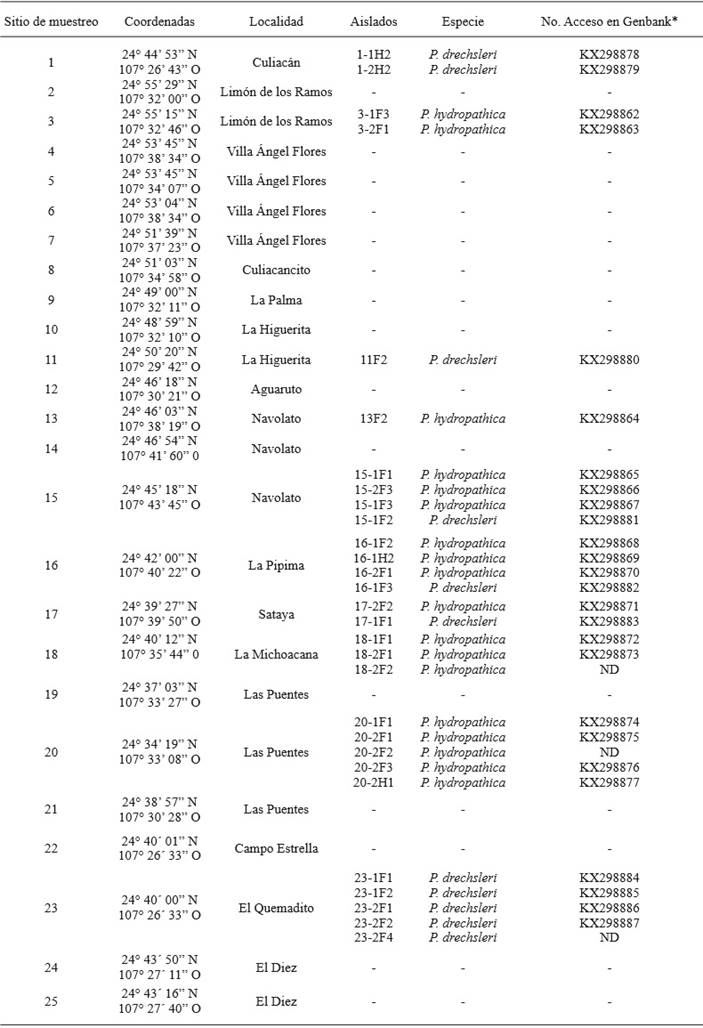
A total of 29 isolates belonging to the genus Phytophthora were obtained from infection sites in pear fruits and azalea leaves. Out of the 25 sampling points, Phytophthora isolations were obtained from only 10 (Table 1). According to their morphological characteristics, isolates were classified into 2 groups: the first group of 18 isolates that produced sporangia with out papilla, of varying shapes, from oval or globular to obpyriforme with a size of 45.8-57.4 x 32.9-36.9 µm (Figure 3A), persistent, with both internal and external proliferations and long and simple sporangiophores; the isolates produced terminal chlamydospores (Figure 3D) and hyphal swellings of irregular shapes in chains and/or in groups (Figure 3C).
These 18 isolates were heterotallic and formed plerotic oospores with an average diameter of 24.9 µm, and oogonios of 27.6 µm, with amphyginousanteridia (Figure 3B). The sexual reproduction of the isolates took place only in reference isolation P. nicotianae A2, indicating that the isolates belong to the compatibility type A1. The maximum growth temperature of the isolates was 40 °C.
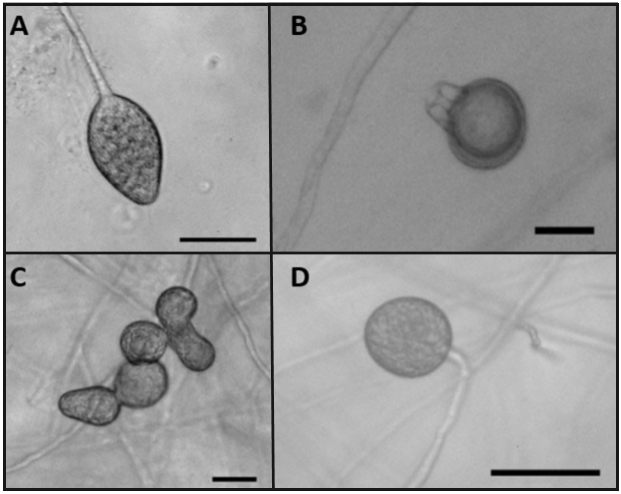
Figure 3. Morphological structure of isolate 3-1F3 of the species P. hydropathica. (A) Non-papillated sporangia, oval-shaped (B) Pleroticoospore with anmphyginousantheridium. (C) Irregular hyphal swellings in groups. (D) Spherical and terminal chlamydospore. Bar = 20 µm.
These morphological characteristics coincide with those for the species Phytophthora hydropathica described in the key proposed by Martin et al. (2012), as well as the description by Hong et al. (2010). It is worth mentioning that these two descriptions of P. hydropathica mention that the species Phytophthora parsiana presents very similar morphological characteristics. Both researchers set them apart based on the maximum temperature growth, since P. hydropathicacan grow in temperatures of up to 40 °C, whereas P. parsiana does not grow in temperatures above 37 °C. Also, Martin et al. (2012) mention that P. parsiana does not present chlamidospores, and P. hydropathica does.
The 18 isolates with chlamidosporeshad a compatibility type A1 when confronted with A1 and A2 strains of P. nicotianae. This coincides with reports by Hong et al. (2010), who report the compatibility type A1in P. hydropathica, therefore this species may not be reproducing sexually, and may survive by means of chlamidospores. According to the morphological characteristics observed, the 18 isolatioisolatesns with nonpapillate sporangia belong to the species Phytophthora hydropathica.
The second group, composed of 11 isolates, share morphological characteristics that consist of a regular petaloid growth pattern in PDA culture medium, non-papillat esporangia with globular and oval shapes, mainly with conical bases and sizes of 25.3-42.7 x 18.702-31.5 µm (Figure 4A), persistent, with internal an external proliferations (Figure 4B) and with simple sporangiophores; this group of isolates produced hyphal swellings (Figure 4C) and no chlamydospores. The isolates in this group were heterothallic,and their compatibility type was A1, forming pleroticoospores with an average diameter of 25.2 µm and 12.25 µm antheridia (Figure 4D). In Michoacán, Mexico the two mating types of mating were found in this species (Mora, 2013), therefore it is likely that if a broader sampling were to be carried out, the type of compatibility A2 could be found. Another characteristic of this group of isolates is the maximum growth temperature, which fluctuates between 35 and 37 °C.
Figure 4. Morphological structures of isolate 17-1F1 of the species P. drechsleri. (A) Non-papillated sporangia, oval-shaped and with a conical base. Bar = 50 µm. (B) extended proliferation of the sporangium. Bar = 50 µm. (C) hyphal swellings. Bar = 50 µm. (D) pleroticoospore with amphyginousantheridium. Bar = 20 µm.
When comparing these characteristics with the taxonomical keys by Erwin and Ribeiro (1996), and Martin et al. (2012), they correspond to the species Phytophthora drechsleri. It is important to point out that both keys mention a relationship between this species and P. cryptogea, although there are also differences between species. One of the most important is the maximum growth temperature, since there are reports indicating that P. cryptogea can not grow in temperatures of 35 °C or above (Perlerou et al., 2010; Kurtbetli, 2014), whereas reports for P. drechsleri indicate it can grow in temperatures of up to 37 °C. There are other differences between both, such as the size of oospores and antheridia: P. drechsleri produces oospores sized approximately 25.6 µm and the oospores from P. cryptogea are smaller, with sizes of around 22 µm. The antheridia comprise a similar case, in which P. cryptogea presents largerantheridia, reaching up to 19 µm in length, while antheridia in P. drechsleri only reach 14 µm. According to the morphological characteristics and the taxonomic keys, we can infer that the group of 11 isolates belongs to the species P. drechsleri.
The PCR reactions with primers DC6-ITS4 with isolates belonging to the species P. hydropathica amplified a product of approximately 1300 pb (Data not shown) and with the primers ELONGF1-ELONGR1, a product of approximately 960 pb (Figure 5). The consensus sequences showed a similarity of 96-98 % with the species Phytophthora hydropathica (code PD 00342 ITS) in the database specialized in Phytophthora (Phytophthora db.org) for primers ITS, and a similarity of 99-100 % with the species of the type P. hydropathica reported in NCBI for both types of primers (EU583793 and GQ260061). This analysis confirms that these isolations belong to the species Phytophthora hydropathica.
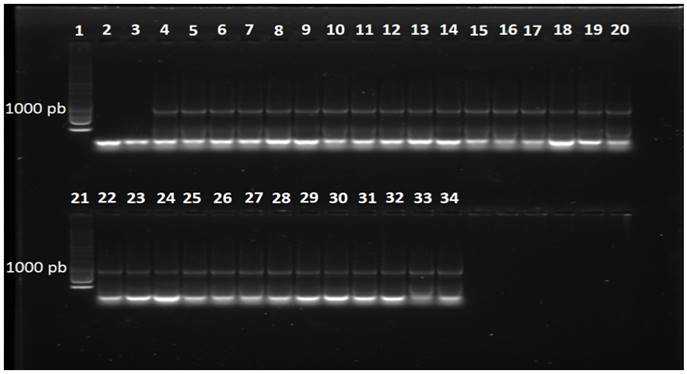
Figure 5. Fragment of DNA amplified with primers ELONGF1/ELONGR1 in isolates obtained from irrigation water. Line 1, 1 Kb molecular marker; Line2, water; Line3, negative control (C. gloeosporioides); Line4, positive control (P. capsici); Lines5-15, isolates of the species P. drechsleri; Lines16-20, isolates of the speciesP. hydropathica; Line21, 1 Kb molecular marker; Lines22-34, isolates of the species P. hydropathica.
P. hydropathica has been frequently analyzed from superficial waters (Bienaflp and Balci, 2014; Vitale et al., 2014; Copes et al., 2015), mainly in the hottest months (Copes et al., 2015) tolerating temperatures up to 40 °C (Hong et al., 2010). It has also been isolated from diseased Japanese andromeda (Pieris japonica) plants (Bienalpf y Balci, 2014) in greenhouses and water used in the irrigation of greenhouses (Hulveyet al., 2010), and from ornamental Viburnumtinus plants with symptoms of shoot dieback (Vitale et al., 2014).
In the P. drechsleri isolation, the reactions of PCR with primers DC6-ITS4 and ELONGF1-ELONGR1 amplified a product of approximately 1300pb (Data not shown) and 970 pb (Figure 5), respectively. The consensus sequences showed a homology of 97 % with isolates reported in the NCBI for the species Phytophthora drechsleri (GU111627 and AY659550), thus confirming the identity of these isolates. P. drechsleri has been found in irrigation water at different depths. For example, Bush et al. (2003) found this species on the surface of irrigation water and at depths of 1 and 1.5 m.
Two species of Phytophthora were isolated from the surface of 10 irrigation channels in the Culiacán Valley. In the channels where Phytophthora was found, P. hydropathical was found in only 40 %, in 30 % only P.drechsleri, and in 30 %, both species (Figure 6). P. hydropathica is considered a native species to river ecosystems (Hong et al., 2010), therefore it is very common to find, in this type of studies, a high frequency of isolates of this species (Bienapfl and Balci, 2014; Brazee et al. 2016).
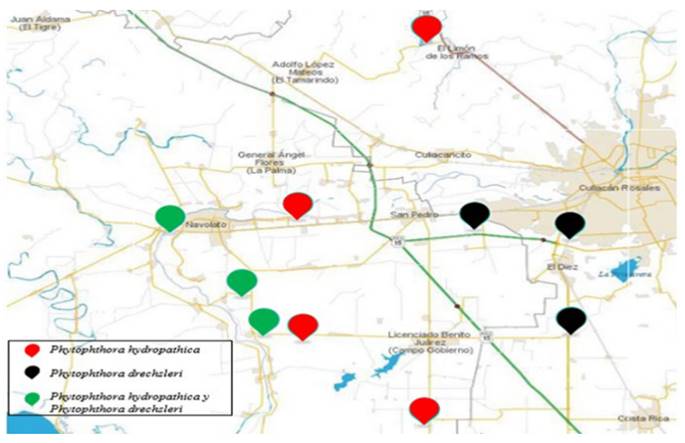
Figure 6. Distribution of P. hydropathica and P. drechsleri in irrigation channels in the Culiacán Valley.
The sequences obtained with primers ELONGF1-ELONGR1 that correspond to the sequences of the gene that codifies for elongation factor 1-α in both species were deposited in the NCBI database (Table 1).
In the pathogenicity tests, representative P. hydropathica isolates 13F2, 15-1F1, 3-2F1, 18-2F1, 16-2F1, 20-2F1, 15-2F3, 3-1F3, 16-1F2, and 16-1H2 caused visible necrosis symptoms 48 h after inoculation in tomato leaflets and chilli pepper leaves (Table 2; Figure 7), as opposed to the leaves used as a control, where there were no symptoms present. The isolations were more aggressive on chilli leaves, causing necrosis on the entire leaf (45 mm), 120 h after inoculation, while in tomato leaves, the largest lesion measured 26 mm (Table 2). Hong et al. (2010) mention chilli pepper, tomato, and cucumber plants as possible hosts, which was confirmed in this study for chilli and tomato. These studies show that P. hydropathica is a threat for tomato and chilli crops, which are extremely important in this region. Similar studies (Bienapfl and Balci, 2014; Brazee et al., 2016), mention that although P. hydropathica rarely has been found to have a relation to plant diseases, it does have the potential to cause diseases.
Table 2. Length of the lesion caused by P. hydropathica and P. drechsleri in chili pepper leaves and tomato leaflets.
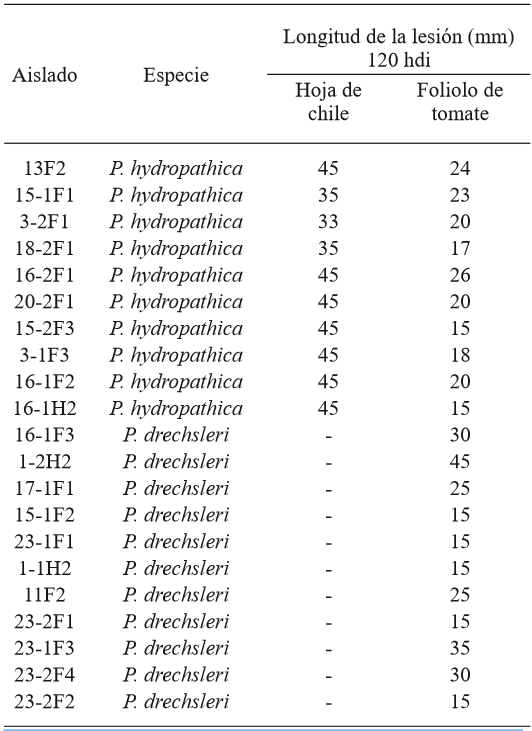
hdi= hours after inoculation; 45 mm= complete leaf.
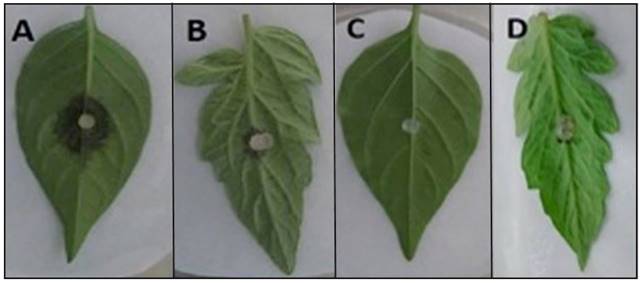
Figure 7. Pathogenicity of P. hydropathica 3-1F3 in chili pepper leaves and tomato leaflets 48 h after inoculation. (A) Inoculated chili leaf. (B) Inoculated tomato leaflet. (C) and (D) Control chili leaf and tomato leaflet.
Isolates16-1F3, 1-2H2, 17-1F1, 15-1F2, 23-1F1, 1-1H2, 11F2, 23-2F1, 23-1F3, 23-2F4 y 23-2F2 of the species P. drechsleri only infected tomato leaf lets (Table 2; Figure 8), in which symptoms of necrosis were observed approximately 48 h after inoculation. Lesions in tomato leaf lets measured up to 45 mm after 120 h, while chilli leaves showed no apparent symptoms, possibly due to the hybrid used for the bioassay may be tolerant to the isolates used. Erwin and Ribeiro (1996) mention that P. drechsleri has been reported as the causal agent of fruit rot in tomato and chili. In Mexico, P. drechsleri has been reported to infects a fflower, chili pepper, gerbera, lettuce, poinsettia, and tomato (Mills et al., 1991; Romero-Cova and Solis-Aragon, 1996; Fernandez-Pavía et al., 2013).

Figure 8. Pathogenicity of P. drechsleri 1-2H2 in tomato leaflets 48 h after inoculation. (A) Inoculated tomato leaflet. (B) Control tomato leaflet. (C) Inoculated chili leaf. (D) Control chili leaf.
This first study in Mexico in the irrigation channels in the CuliacánValley shows the presence of P. hydropathica and P. drechsleri and highlights the importance of irrigation water as a way of spreading both pathogens. At the same time, it lays the foundations for future studies, such as knowing the population dynamics through out the agricultural cycle, determining whether these species only use irrigation channels as a way of dissemination or if they complete their life cycle in these bodies of water, as well as to determining the presence of P. hydropathica and P. drechsleri in natural infections of horticulturally important plants in the region. This is the first report on Phytophthora hydropathica in irrigation channels in Mexico. These results indicate that it is necessary to continue research on natural environments, whether in superficial waters or in soils, to know the diverse species of Phytophthora present in Mexico, and therefore consider the potential risk these microorganisms represent for Mexican agriculture.
Acknowledgements
To CONACYT for funding Brando Álvarez Rodriguez’s studies.
REFERENCES
Bienapfl JC and Balci Y. 2014. Movement of Phytophthora spp. in Maryland ́s trade. Plant Disease 98:134-144. http://dx.doi.org/10.1094/PDIS-06-13-0662-RE [ Links ]
Blair JE, Coffey M, Park S, Geiser DM and Kang S. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology 45:266-277. http://dx.doi.org/10.1016/j.fgb.2007.10.010 [ Links ]
Bonants P, Hagenaar de Weerdt M, van Gent-Pelzer M, Lacourt, Cooke D and Duncan M. 1997. Detection and identification of Phytophthora fragariae Hickman by the polymerase chain reaction. European Journal of Plant Pathology 103:345-355. Disponible en línea: http://link.springer.com/article/10.1023%2FA%3A1008640227432 [ Links ]
Brazee NJ, Wick RL and Hulvey JP. 2016. Phytophthora species recovered from the Connecticut River Valley in Massachusetts, USA. Mycologia 108:6-19. http://dx.doi.org/10.3852/15-038 [ Links ]
Bush EA, Hong CX and Stromberg EL. 2003. Fluctuations of Phytophthora and Pythium spp. in components of a recycling irrigation system. Plant Disease 87:1500-1506. http://dx.doi.org/10.1094/PDIS.2003.87.12.1500 [ Links ]
Copes WE, Yang X and Hong CX. 2015. Phytophthora species recovered from irrigation reservoirs in Mississippi and Alabama nurseries and pathogenicity of three new species. Plant Disease 99:1390-1395. http://dx.doi.org/10.1094/PDIS-11-14-1197-RE [ Links ]
Erwin DC and Ribeiro OK. 1996. Phytophthora diseases worldwide. First edition, APS press, St. Paul, MN. 562 p. [ Links ]
Fernandez-Pavía SP, Díaz-Celaya M and Rodríguez-Alvarado G. 2013. Phytophthora in Mexico. Pp: 215-221. In: Lamour K. (ed.). Phytophthora: A global perspective. CAB International. USA. 257 p. [ Links ]
Fernández-Pavía SP, Gregorio-Cipriano R, Rodríguez-Alvarado G, Fernández-Pavía YL, Mondragón-Flores A, Gómez-Dorantes N, Lozoya-Saldaña H, Rodríguez-Fernandez R y Herrera-Camacho J. 2015. Enfermedades de especies vegetales en México. Universidad Michoacana de San Nicolás de Hidalgo. Primera edición. Morelia, México. 425 p. [ Links ]
Gallegly ME and Hong CX. 2008. Phytophthora: identifying species by morphology and DNA fingerprints. Primera edición. APS Press. St Paul, MN, USA. 168 p. [ Links ]
Ghimire SR, Richardson PA, Moorman GW, Lea-Cox JD, Ross DS and Hong CX. 2009. An in-situ baiting bioassay for detecting Phytophthora species in irrigation runoff containment basins. Plant Pathology 58:577-583. http://dx.doi.org/10.1111/j.1365-3059.2008.02016.x [ Links ]
Ghimire SR, Richardson PA, Kong PHJ, Lea-Cox JD, Ross DS, Moorman GW and Hong CX. 2011. Distribution and diversity of Phytophthora species in nursery irrigation reservoir adopting water recycling system during winter months. Journal of Phytopathology 159:713-719. http://dx.doi.org/10.1111/j.1439-0434.2011.01831.x [ Links ]
Grünwald NJ, Martin FN, Larsen MM, Sullivan CM, Press CM, Coffey MD, et al. 2011. Phytophthora-ID.org: A sequence-based Phytophthora identification tool. Plant Disease 95:337-342. http://dx.doi.org/10.1094/PDIS-08-10-0609 [ Links ]
Hajebrahimi S and Banihashemi Z. 2011. Host range of Phytophthora parsiana: a new high temperature pathogen of woody plants. Phytopathologia Mediterranea 50:159-165. http://dx.doi.org/10.14601/Phytopathol_Mediterr-3055 [ Links ]
Hardham AR. 2005. Phytophthora cinnamomi. Molecular Plant Pathology 6:589-604. http://dx.doi.org/10.1111/J.1364-3703.2005.00308.X [ Links ]
Hong CX, Gallegly ME, Richardson PA, Kong P, Moorman GW, Lea-Cox JD and Ross DS. 2010. Phytophthora hydropathica, a new pathogen identified from irrigation water, Rhododendron catawbiense and Kalmia latifolia. Plant Pathology 59:913-921. http://dx.doi.org/10.1111/j.1365-3059.2010.02323.x [ Links ]
Hong CX and Moorman GW. 2005. Plant pathogens in irrigation water: Challenges and Opportunities. Critical Reviews in Plant Science 24:189-208. Disponible en línea: http://www.nurserycropscience.info/water/pathogens/technical-pubs/hong-and-moorman-2005.pdf/view [ Links ]
Hüberli D, Hardy GE St J, White D, Williams N and Burgess TI. 2013 Fishing for Phytophthora from Western Australia ́s waterways: a distribution and diversity survey. Australasian Plant Pathology 42:251-260. http://dx.doi.org/10.1007/s13313-012-0195-6 [ Links ]
Hulvey J, Gobena D, Finley L and Lamour K. 2010. Co-occurrence and genotypic distribution of Phytophthora species recovered from watersheds and plant nurseries of eastern Tennessee. Mycologia 102:1127-1133. http://dx.doi.org/10.3852/09-221 [ Links ]
Jeffers SN and Martin SB. 1986. Comparison of two media selective for Phytophthora and Pythium species. Plant Disease 70:1038-1043. Disponible en línea: https://www.apsnet.org/publications/PlantDisease/BackIssues/Documents/1986Articles/PlantDisease70n11_1038.pdf [ Links ]
Kroon LPNM, Bakker FT, van den Bosh GBM, Bonants PJM and Flier WG. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology 41:766-782. http://dx.doi.org/10.1016/j.fgb.2004.03.007 [ Links ]
Kurtbetli I. 2014. Involvement of Phytophthora cryptogea in sweet cherry decline in Turkey. Phytoparasitica 42:627-630. http://dx.doi.org/10.1007/s12600-014-0403-8 [ Links ]
Martin FN, Abad ZG, Balci Y and Ivors K. 2012. Identification and detection of Phytophthora: Reviewing our progress, identifying our needs. Plant Disease 96:1080-1103. http://dx.doi.org/10.1094/PDIS-12-11-1036-FE [ Links ]
Mills SD, Foerster H and Coffey MD. 1991. Taxonomic structure of Phytophthora cryptogea and P. drechsleri based on isozyme and mitochondrial DNA analyses. Mycological Research. 95:31-48. http://dx.doi.org/10.1016/S0953-7562(09)81359-2 [ Links ]
Mitchell DJ and Kannwischer-Mitchell ME. 1992. Phytophthora. Pp: 31-38. In: Singleton LE, Mihail JD and Riush CM. (eds.). Methods for research on soilborne phytopathogenic fungi. American Phytopathological Society, St, Paul, MN. 266 p. [ Links ]
Mora DAN. 2013. Variabilidad genética de aislados de P. capsici y Phytophthora drechsleri de plantas ornamentales. Tesis de Maestría. Universidad Michoacana de San Nicolás de Hidalgo. Morelia, Michoacán. 112 p. [ Links ]
Orlikowski LB, Ptaszek ATM and Orlikowska T. 2012. Relationship between source of water, occurrence and pathogenicity of Phytophthora plurivora. Acta Mycologica 47:3-9. Disponible en línea: https://pbsociety.org.pl/journals/index.php/am/article/viewFile/am.2012.001/2195 [ Links ]
Perlerou C, Tziros G, Vettraino AM and Diamandis S. 2010. Phytophthora cryptogea causing ink disease of Castanea sativa newly reported in Greece. Plant Pathology 59:799. http://dx.doi.org/10.1111/j.1365-3059.2010.02268.x [ Links ]
Reeser PW, Sutton W, Hansen EM, Remigi P and Adams GC. 2011. Phytophthora species in forest streams in Oregon and Alaska. Mycologia 103:22-35. http://dx.doi.org/10.3852/10-013 [ Links ]
Romero-Cova S and Solis-Aragon G. 1996. New hosts of some species of the genus Phytophthora (Phycomycetes: Pythiaceae) in Mexico. Agrociencia 30:241-247. [ Links ]
Sutton W, Hansen EM, Reeser PW and Kanaskie A. 2009. Stream monitoring for detection of Phytophthora ramorum in Oregon tanoak forest. Plant Disease 93:1182-1186. http://dx.doi.org/10.1094/PDIS-93-11-1182 [ Links ]
Tyler BM. 2007. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Molecular Plant Pathology 8:1-8. http://dx.doi.org/10.1111/j.1364-3703.2006.00373.x. [ Links ]
Vitale S, Luongo L, Galli M and Belisario A. 2014. First report of Phytophthora hydropathica causing wilting and shoot dieback on Viburnum in Italy. Plant Disease 98:1582. http://dx.doi.org/10.1094/PDIS-03-14-0308-PDN [ Links ]
White TJ, Bruns T, Lee S and Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pp: 315-322. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds.) PCR protocols: A guide to methods and applications. Academic Press, San Diego, U.S.A. 461 p. [ Links ]
Yamak F, Peever TL, Grove GG and Boal RJ. 2002. Occurrence and identification of Phytophthora spp. pathogenic to pear fruit in irrigation water in the Wenatchee River valley of Washington State. Phytopathology 92:1210-1217. http://dx.doi.org/10.1094/PHYTO.2002.92.11.1210. [ Links ]
Zappia RE, Hüberli D, Hardy GE St. J and Bayliss KL. 2014. Fungi and oomycetes in open irrigation systems: knowledge gaps and biosecurity implications. Plant Pathology 63:961-972. http://dx.doi.org/10.1111/ppa.12223 [ Links ]
Zelaya-Molina L, Ortega M and Dorrance A. 2011. Easy and efficient protocol for oomycete DNA extraction suitable for population genetic analysis. Biotechnology Letters 33:715-720. http://dx.doi.org/10.1007/s10529-010-0478-3 [ Links ]
Received: June 13, 2016; Accepted: September 27, 2016











 text in
text in 




