A great diversity of pathogenic microorganisms such as Oomycetes, fungi, bacteria, nematodes, among others, can cause diseases in crop plants, being a serious problem causing economic losses worldwide. Therefore, the problems and limitations of the control of plant diseases have been handled by using chemical pesticides. Furthermore, its inappropriate use has been linked to several problems such as environmental pollution, food residues, intoxication risk when they are managed without adequate protection equipment, also resistant development of pathogens to the synthetic fungicides and the loss of the natural biodiversity in agroecosystems (Harris et al., 2001; Fisher et al., 2012). This problematic has conducted to the search of new alternatives for managing plant diseases that are ecologically and economically viable, with minimal impact on the environment.
An alternative to chemical control of phytopathogenic fungi is its suppression by use of biological control agents with emphasis in the use of native antagonists (Zeilinger and Omann, 2007; Vinale et al., 2008), being a tool that does not affect the environment. Consideration has been given to the possibility of controlling pathogens by the use of several microorganisms with antagonistic activity such as fungi, bacteria, and actinomycetes. However, the first two have been the most used, mainly Trichoderma and Bacillus genera. The most used bacteria specie has been B. subtilis, however there are other antagonist species such as B. methylotrophicus and B. amyloliquefaciens which have been poorly studied, but have been considered as efficient colonizers that are widely spread in different habitats, due to its capacity to form spores, grow in a wide range of temperatures, antibiotic production that inhibit the growth of phytopathogens, besides being plant growth promoters (Leelasuphakul et al., 2008). Both, antagonistic fungi and bacteria, constitute the majority of the microbial population in soil that is the main natural reservoir, where the equilibrium between the diversity of microorganism is dynamic, contributing in reducing diseases in crop plants and uncultivated plants (Gajera and Vakharia, 2010; Singh and Islam, 2010). This ecological balance is supported by different complex interactions to inhibit the growth of phytopathogenic fungi through several modes of action such as: direct competition for nutrients and space, direct mycoparasitism, production of antibiotics by the excretion of toxic compounds like cytokinins and auxins, inactivation of pathogenic enzymes, also can be compatible with management practices, including the use of pesticides, production of organic and inorganic volatile compounds. Besides its fast growth, are easily adaptable, induces systemic resistance in host plants to pathogens, and contributes to a better assimilation of nutrients (Ezziyyani et al., 2004; Harman, 2006; Leelasuphakul et al., 2008; Vinale et al., 2008; Arguelles, 2009; Correa et al., 2009).
However, antagonistic microorganisms populations have been reduced by anthropogenic activities, especially by chemical pesticides used in the crops management (Gajera and Vakharia, 2010). Microbial control agents are commonly used for the management of bacterial and fungal diseases in crops and it is becoming more widely used, with a possible reduction in the use of synthetic pesticides. These microbial agents are generally identified based on their microscopic morphological characters. Although microorganisms can be identified based on morphologic characters, molecular techniques are nowadays widely used and in general more acceptable tools for identification that offer quick and reliable information for the study of identity (Gajera and Vakharia, 2010). Therefore the aims of this study were to identify taxa and to evaluate the antagonistic activity in vitro of two Bacillus isolates and two Trichoderma asperellum isolates, against five common pathogenic fungi.
Experimental site and biological samples
The experiment was performed in vitro, in the Laboratory of Patología Vegetal y Control Biológico of the Centro de Investigación en Alimentación y Desarrollo, A.C., Campus Cuauhtémoc, Chihuahua, México. Microorganisms used, were obtained from the microorganisms ceparium of the same Campus. Pathogens were isolated from fruits, diseased tissue of apple trees and soil samples near to rhizosphere, collected from different apple orchards and geographic regions of Chihuahua, México, in different periods. Also antagonists were obtained from soil samples near to rhizosphere of apple trees in the same geographic regions and were selected by their antagonistic capacity in vitro against Oomycetes (Rios-Velasco et al., 2014). The fungi were active into potato-dextrose-agar (PDA, BioxonTM Becton Dickinson de México, S.A. de C.V.) and incubated at 26 °C in an environmental chamber (Precision Scientific, Model 6LM, Winchester, VA, USA) for 5-10 d.
Morphological and molecular identification
Nine microorganisms: five phytopathogenic fungi, two antagonistic fungi and two antagonistic bacteria isolates, used in this study were identified to genus with the help of taxonomic keys according to their morphological characters as viewed by an optical microscope (Carl Zeiss, Jena, Germany) (Dugan, 2006; Watanabe, 2010). Subsequently, were molecularly identified in the Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, CINVESTAV, Campus Irapuato, Guanajuato, México.
To identify the fungal isolates based on their molecular characters, their genomic DNA (gDNA) was extracted. Thus, an explant of each purified fungus was placed and cultured in a Petri dish with PDA covered with a sterile cellophane to facilitate the collection of the fungal biomass and incubated at 26 °C for 7 d. Mycelium was collected in a porcelain mortar together with a buffer [200 mM Tris-HCl at pH = 8, 250 Mm NaCl, 25 mM EDTA, 0.5 % SDS] for gDNA extraction filamentous fungi at 70 °C, frozen with liquid nitrogen and macerated following the protocol described by Raeder and Broda (1985). The gDNA obtained from fungi was examined by electrophoresis in an 1 % agarose gel, which was used to amplify the 18S rDNA gene and the Internal Transcribed Spacer (ITS), using the universal primers ITS5 (5'-GGAAGTAAAAGTCGTAACAAGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3') where the expected fragments were about 600 to 710 bp (White et al., 1990). The amplification conditions were divided into various steps: an initial step of denaturalization at 94 °C for 5 min, a second step comprising 30 cycles including denaturalization at 94 °C of 30 s, an alignment step at 60 °C for 30 s, a step of extension at 72 °C for 45 s, and when the cycles were completed, a final extension at 72 °C for 10 min.
For the antagonistic bacteria, an explant of the purified colony of 72 h growth was taken for the gDNA extraction described by Heddi et al. (1999). The gDNA obtained was examined by electrophoresis in an 1 % agarose gel, which was used to amplify the gene of the 16S of the rDNA by PCR, using the universal primers EU(F) (5'-AGAGTTTGATCATGGCTCAG-3') and EU(R) (5'-TACCTTGTTACGACTTCACC-3'). The amplification conditions were divided into various steps: an initial of denaturalization at 94 °C for 5 min, a second step comprising 35 cycles including denaturalization at 94 °C for 30 s, an alignment step at 53 °C for 45 s, a step of extension at 72 °C for 45 s, and when the cycles were completed, a final extension at 72 °C for 10 min (Heddi et al., 1999).
The PCR products of both microorganisms' taxa were examined by electrophoresis in an 1 % agarose gel. Subsequently, were sequenced by the company Macrogen (Rockville, Maryland, USA). Sequences obtained were compared against the NCBI database using the BLAST algorithm (Altschul et al., 1990) to verify the percent identity and they corresponded to the identified species. The nucleotide sequences obtained, have not been deposited in the NCBI.
Inoculum preparation
The inoculum was prepared by a suspension of fungal spores and bacteria, respectively, in sterile peptone-water containing sodium chloride at 0.85 % and peptone at 0.1 % into 2 mL micro tubes. The concentration of spores was estimated using a Neubauer chamber (Neubauer Improved bright-line, Marienfield, Germany) and bacteria were quantified by technique of serial dilutions and plate colony count. Both suspensions were stored at 0 °C until use. The concentration of the suspensions where the filter paper circles were immerse for the evaluation of antagonistic activity in vitro, ranged from 1.0 × 107 to 9.5 × 107 fungi spores or CFU-bacteria per milliliter.
In vitro evaluation of antagonism
In vitro confrontations of T. asperellum against pathogenic fungi were performed using the dual culture technique (Petri dishes, 90 × 15 mm) with PDA, that consisted in placing in one side of the dish a filter paper circle of 6 mm in diameter, impregnated with a suspension of conidia and mycelium as the inoculum of pathogens and in the opposite side of the Petri dish, another filter paper circle with mycelium and/or conidia of the antagonist was placed. Ten replicates for each evaluation were performed by triplicate. Also ten controls (Petri dishes) by triplicate were considered, both phytopathogens as antagonists, grown separately. Petri dishes inoculated were incubated at 28 °C for 15 d in an environmental chamber (Precision Scientific, Model 6LM, Winchester, USA). The radial growth of the fungal colonies was measured every 24 h for both microorganisms pathogens and antagonists. The inhibition halo, between phytopathogen and antagonist colonies in confrontation, was measured to the cardinal points of the Petri dish at 14 d post-incubation (Aquino-Martínez et al., 2008).
The time when pathogen-antagonist colonies contacted, inhibition of radial growth of the colony, and type of antagonism according to Bell's scale (Bell et al., 1982) were evaluated. Where the antagonistic activity was measured by five levels, being the level one of the scale when the antagonist overgrown completely the pathogen and fills the culture surface and the level five was when the pathogen overgrown completely to the antagonist and fills the culture surface.
The antagonism of T. asperellum was evaluated registering the following variables: radial growth of the antagonist (RGA), radial growth of the pathogen (RGP), and percentage of radial growth inhibition (PRGI). The PRGI was determined at sixth day post-inoculation using the formula proposed by Ezziyyani et al. (2004), PRGI = [(R1 - R2) / R1] × 100, where R1 was the radial growth of the control colony (pathogen) and R2 was the radial growth of the pathogen colony in the in vitro confrontation.
In vitro confrontation of Bacillus species against pathogens were performed by placing one filter paper circle of 6 mm in diameter of pathogen in the center of Petri dish containing PDA, whereas the bacteria were inoculated on the cardinal points of the Petri dish, using filter paper circles of 6 mm in diameter with the inoculum. Ten replicates (Petri dishes) by triplicate were performed for each bacterium-fungus in vitro evaluation. Each of the control treatments of phytopathogenic fungi was cultured separately and incubated at 28 °C for 14 d. The radial growth of the pathogen in confrontation towards the antagonist colony was measured daily and in controls the radial growth from the center of the Petri dish toward the cardinal points was measured.
Statistical analysis
Both bioassays were performed in triplicate, with ten Petri dishes per replicate, for each bacterium-fungus in vitro evaluation, using a completely randomized design, with five treatments: two antagonistic fungal isolates and two antagonistic bacteria isolates, confronted against five pathogens, besides 1 control, where each treatment was an antagonist microorganism with 30 Petri dishes (i.e., a total of 600). Data of PRGI and inhibition halo were analyzed using the Statistical Analysis System version 9.0 (SAS, 2002) for balanced analysis of variance (ANOVA) and means were separated by Tukey's test (P ≤ 0.05).
Pathogens and antagonists identified
The phytopathogens fungi identified morphologically and from the sequences of the PCR products were: Fusarium oxysporum strain LCF32, Botrytis cinerea strain G409, Penicillium crustosum strain 06CK005, Aspergillus nidulans strain UOA/HCPF, Alternaria alternata strain HMY 2-1, which had a 99-100 % of identity and a maximum similarity with the molecular scores and taxonomic keys excepting P. crustosum, that showed a 78 % of identity, corresponding to each strain and according to the sequences available in the GenBank database (NCBI), obtained by the BLAST algorithm (Altschul et al., 1990). Also sequencing data showed that the fungal and bacterial antagonistic isolates identified were: Trichoderma asperellum TC1 strain LAHC-FFPK-M16 and T. asperellum TC2 strain BHU-BOT-RYRL16, Bacillus methylotrophicus strain IS04, Bacillus amyloliquefaciens strain Abk-2, had 99-100 % identity and maximum scores when compared with GenBank sequence, obtained from the BLAST algorithm NCBI database (Altschul et al., 1990).
Antagonistic activity
In vitro . Radial growth of the five phytopathogenic fungi tested, was significantly reduced when evaluated in vitro against T. asperellum TC1 and TC2, where both antagonists fungi showed a significant efficiency to inhibit the growth of phytopathogens fungi at 6-8 d post-evaluation (Table 1, Figure 1). Petri dishes of the controls of both T. asperellum isolates were completely covered by the growth of the fungi at the third day post-inoculation. Meanwhile at sixth day post-inoculation, both TC1 and TC2 isolates, came in contact with F. oxysporum, B. cinerea, P. crustosum, and A. alternata except with A. nidulans. The highest PRGI rates were shown in B. fabae with both T. asperellum TC1 and TC2 isolates with 71.6 and 69.8 %, respectively (Table 1), while the lowest rates were observed in P. crustosum and F. oxysporum with 52.3 and 43.3 %, respectively (Table 1).
Table 1 In vitro radial growth of phytopathogenic fungi, when inhibited by Bacillus species 14 d post-inoculation and by Trichoderma species at sixth day of incubation.
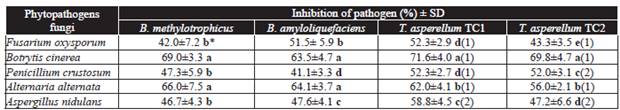
*Letters in the same column indicate significant differences in the treatments where the means followed by the same letter are not significantly different (ANOVA) according to Tukey's test (P ≤ 0.05).
Antagonistic activity was measured according to scale of Bell et al. (1982) where: (1) antagonist (Trichoderma) overgrown completely on pathogen's colony and completely covered medium's surface and (2) antagonist (Tricoderma) two thirds of medium surface overgrown. SD = Standard deviation.
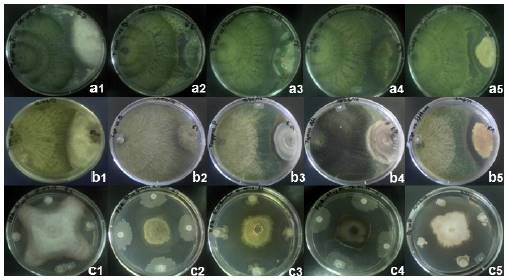
Figure 1 Mycoparasitism of Trichoderma asperellum TC1 (a1-a5) and Trichoderma asperellum TC2 (b1-b5), and inhibition halos of Bacillus amyloliquefaciens (c1-c5) when confronted in vitro against Fusarium oxysporum, Botrytis cinerea, Penicillium crustosum, Alternaria alternata, and Aspergillus nidulans, respectively, at 14 d post-inoculation.
In vitro evaluations against A. nidulans, inhibition halos of 2.6 and 4.5 mm were observed with T. asperellum TC1 and TC2 isolates, respectively (Table 2, Fig. 1), this may be due to the production of volatile compounds that inhibited the growth of Trichoderma spp., even so, both isolates were efficient to reduce the phytopathogenic fungi growth at sixth day post-inoculation (Duffy et al., 2003; Harman, 2006; Zeilinger and Omann, 2007; Vinale et al., 2008). To respect, Cooney et al. (2001), mentioned that T. harzianum produces the antibiotic 6-pentyl-α-pyrone, which has a dual effect in inhibiting the growth of the pathogen and can also regulate genes for trichotecenes biosynthesis and micotoxines with broad spectrum antimicrobial activity. Also, Gajera and Vakharia (2010) reported a highest inhibition of the growth of A. niger, caused by T. viride and T. harzianum during a study performed in vitro. Besides having the ability to produce wall cell degrading enzymes such as chitinase, β-1,3 glucanase, and protease in the culture medium during the in vitro antagonism. On the other hand, the antagonist or their purified antibiotics, can reduce the production of deoxynivalenol by >80 % and antagonistic genes are up-regulated by the presence of the pathogen (Duffy et al., 2003). Therefore, the efficiency of Trichoderma spp. to inhibit phytopathogens fungi may have been due to their competition for space and nutrients, antibiotic production (volatile and non-volatile compounds), mycoparasitism and inactivation of the pathogen's enzymes (Ezziyyani et al., 2004).
Table 2 Inhibition halo between colonies of phytopathogenic fungi and the antagonistic Bacillus species 14 d post-inoculation and Trichoderma asperellum isolates at 6 d of incubation.

*Letters in the same column indicate significant differences in the treatments where the means followed by the same letter are not significantly different (ANOVA) according to Tukey's test (P ≤ 0.05). SD = Standard deviation.
In addition, an inhibition halo was observed in T. asperellum TC2 isolate when evaluated in vitro against P. crustosum (Table 2), this might be due to patulin production, a secondary metabolite produced by this phytopathogen (Leelasuphakul et al., 2008).
The growth inhibition of the pathogenic fungi colonies, induced by B. methylotrophicus and B. amyloliquefaciens, was significantly different, where the highest effect was observed in B. cinerea with PRGIs of 69 and 63.5 %, respectively, A. alternata showed PGRIs 66 and 64.1 %, respectively; whereas the lowest growth reductions were registered for F. oxysporum, P. crustosum, and A. nidulans (Table 1). These results are similar to those obtained by Madhaiyan et al. (2010) and Zhang et al. (2012) who found a high capacity of some strains of Bacillus of the same species to inhibit the growth of several phytopathogens fungi, which was due to the production of volatile compounds and antibiotics belonging to the family of iturins and subtilins, that act on the fungi's cell wall. Also, the production of hydrophilic enzymes to break down polysaccharides, nucleic acids and lipids, which are used as an energy source, might have been involved.
Bacillus amyloliquefaciens and B. methylotrophicus induced an inhibition halo of the pathogen colony at 14 d post-incubation (Table 2). Mean values per antagonist showed that B. methylotrophicus had a higher antagonistic activity than B. amyloliquefaciens, since the average of inhibition halo was significantly, of 4.95 to 13.15 mm and 4.15 to 8.45 mm, respectively, in the phytopathogenic fungi colonies (Table 2). The highest inhibition halos produced by B. amyloliquefaciens and B. methylotrophicus were observed with B. cinerea with 8.45 and 13.25 mm, respectively, whereas the lowest rates were shown against F. oxysporum with 4.15 and 4.95 mm, respectively (Table 2). Bacillus methylotrophicus showed the highest antagonistic effect against phytopathogens that may be due to the production of antibiotics and/or volatile organic compounds as hydrogen cyanide which inhibit the growth of the phytopathogenic fungi and exert harmful effects on the in vitro growth of several phytopathogens, these compounds can have direct or indirect effects in the activity of specific fungal enzyme (Wheatley, 2002; Duffy et al., 2003; Correa et al., 2009; Zhang et al., 2012), as also reported by Madhaiyan et al. (2010) and Zhang et al. (2012), who found that strains of B. methylotrophicus have a high antagonistic activity against a wide diversity of phytopathogens fungi.
According to Guillén-Cruz et al. (2006), B. amyloliquefaciens was the most efficient against Phytophthora spp. when evaluated in vitro, in addition to having an antagonistic effect and induce plant growth. To respect, Souto et al. (2004) mentioned that B. amyloliquefaciens strain added peptides and lipopeptides to the culture medium, such as fungicine, iturin, bacillomicine, among others, having antifungal properties when confronted in vitro against phytopathogenic fungi such as Fusarium, Rhizoctonia, and Sclerotinia.
Both antagonistic bacteria and fungi, significantly inhibited the in vitro growth of F. oxysporum, B. cinerea, P. crustosum, A. alternata, and A. nidulans, where B. methylotrophicus and T. asperellum TC1, were the most efficient, which suggests the presence of bioactive compounds. Therefore, can be considered as viable biological control agents. So it is important to conduct more studies to identify those bioactive compounds and how they suppress the growth of the pathogens.











 texto en
texto en 


