Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista mexicana de fitopatología
versão On-line ISSN 2007-8080versão impressa ISSN 0185-3309
Rev. mex. fitopatol vol.33 no.1 Texcoco 2015
Scientific articles
Identification of Meloidogyne species by sequencing of internal transcribed spacer regions of ribosomal DNA of juvenile stages
1Universidad Autónoma de Nuevo León, UANL, Facultad de Ciencias Biológicas, Instituto de Biotecnología, Pedro de Alba s/n, Cd. Universitaria, San Nicolás de los Garza, N.L. México. C.P. 66450 Tel/fax (81) 83294000, ext. 7304, Fax/ext. 6415.
Usually the identification of the Meloidogyne species is based on the morphology of adult females, making it difficult to identify juvenile males and females (J2). Nematodes are considered among the most difficult animals to identify; the use of ribosomal DNA (rDNA) based diagnostic methods have gained acceptance in applications ranging from quarantine determinations to assessments of biodiversity. Nematodes of the genus Meloidogyne are known for their ability to produce physiological changes in the root system of plants and cause losses in the absorption of nutrients. The objective of this study was to determine if the sequencing of internal transcribed spacer (ITS) regions of rDNA can be used as genetic markers for reliable identification of populations of juvenile males and females (J2) for the main species of the genus Meloidogyne. From samples of diseased tomato roots (Solanum lycopersicum L.), larvae of juvenile females and males of Meloidogyne were collected for the DNA extraction. A rDNA region harboring two ITS regions was amplified. For subsequent sequencing, that region was ligated into pGEM(r)-T vector. Analysis with the BLASTn program showed that the gene region identified 99.8 % with a gene sequence belonging to Meloidogyne incognita Kofoid & White, 1919. This result suggests that the ITS regions can be used as a genetic marker in populations for Meloidogyne species identification.
Key words: Meloidogyne incognita ; ITS; Ribosomal DNA (rDNA)
Usualmente, la identificación de especies de Meloidogyne está basada en la morfología de hembras adultas, lo que dificulta la identificación de hembras y machos juveniles (J2). Los nematodos son considerados entre los animales más difíciles de identificar, el uso de marcadores genéticos basados en el ADN ribosomal (ADNr) ha ganado aceptación en aplicaciones que van desde las determinaciones de cuarentena a las evaluaciones de la biodiversidad. Los nematodos del género Meloidogyne son conocidos por su habilidad para producir cambios fisiológicos en el sistema radical de las plantas y causar pérdidas en la absorción de nutrientes. El objetivo de este estudio fue determinar si la secuenciación de regiones espaciadoras transcritas (ITS) internas de ADNr pueden ser usadas como marcadores genéticos para la identificación confiable de poblaciones de hembras y machos juveniles (J2) para las principales especies del género Meloidogyne. De muestras de raíces enfermas de tomate (Solanum lycopersicum L.) se extrajeron larvas de hembras y machos juveniles de Meloidogyne para la extracción de ADN. Se amplificó una región génica del ADNr que contiene dos regiones ITS. Para su posterior secuenciación, dicha región se ligó al vector pGEM(r)-T. El análisis con el programa BLASTn indicó que la región génica presentó una identidad del 99.8 % respecto a una secuencia génica perteneciente a Meloidogyne incognita Kofoid & White, 1919. Tal resultado sugiere que las regiones ITS pueden ser usadas como un marcador genético en poblaciones de Meloidogyne para la identificación de especie.
Palabras clave: Meloidogyne incognit ; ITS ; ADN ribosomal (ADNr)
The nematodes of the Meloidogyne genus are known for their ability to produce physiological changes in the root system of plants and cause losses in the absorption of nutrients, affecting their growth (dwarfism) and their production (Wishart et al., 2002). However, the effects of the nematodes on crops are underestimated by farmers and agricultural technicians due to the non-specific symptoms, that are often mistaken for lack of nutrients, water stress, problems of soil fertility, and with other secondary infections caused by fungi or bacteria, the entry of which is made easier by the nematode. According to Zuckerman et al. (1990), the parasite nematodes of the plants diminish the global agricultural production between 12 and 20%. Even though there are at least 70 nematode species that cause root nodules, taxonomic attention has focused on six of them (Meloidogyne incognita Kofoid & White, 1919, M. javanica Treub, 1885, M. arenaria Neal, 1889, M. chitwoodi Golden et al., 1980, M. fallax Karssen, 1996 y M. hapla Chitwood, 1949) due to the fact that these are typically associated with agronomically important plants (Adam et al., 2007). For the identification of species and populations of Meloidogyne, several methods have been used, although all of them have their limitations (Zijlstra et al., 1995). It is important to determine the composition of species of Meloidogyne in crops in order to implement control strategies that do not involve any chemicals, such as the rotation of resilient cultivars and allelopathic plants (Cenis, 1993; Orui, 1998). The identification of Meloidogyne species is based on the morphology of adult females (Eisenback et al., 1981) and the range of hosts (Williamson et al., 1997), making the identification for young males and females of the second stage (J ) more difficult. The analysis of isoenzymes is also used, where the comparison of patterns of esterase show a great consistency in the division of the four species most widely distributed in the world; however, it does not detect any intraspecific variation among Meloidogyne nematodes (Volvas et al., 2005). The diagnoses based on the DNA provide some attractive solutions for the problems associated with these methods of identification, due to the fact that they are independent from the products expressed in the genome and from the influence of the environment and the stage of the nematodes (Wishart et al., 2002; Powers, 2004). Most DNA analyses directed at nematodes that cause the formation of galls use total DNA (genomic and mitochondrial [mtDNA]) or mtDNA, and are predominantly done through Restriction Fragment Length Polymorphisms (RFLP) (Zijlstra et al. 1995, Oruri, 1998; Powers, 2004). The comparison between sequences of internal transcribed spacer regions (ITS) of ribosomal DNA (rDNA) is used in taxonomy, due to its high level of polymorphism between closely related species and is virtually a constant for one specific species. Furthermore, the ITS regions are lined by retained sequences that facilitate the design of indicators for their expansion by the Polymerase chain reaction (PCR).
The objective of this study is to determine if the internal transcribed spacer regions of rDNA can be used as genetic markers for the reliable identification of young female and male J2 populations don main species of the Meloidogyne genus.
Materials and methods
Biological material
The juvenile J2 of Meloidogyne were obtained using the Baermann funnel technique (Agrios, 2005) of tomato plant roots with gall formations (Solanum lycopersicum), a farm located in San Nicolás de los Garza Nuevo León, provided by the Department of Plant Pathology of the Faculty of Life Sciences at the Autonomous University of Nuevo León, and identified as belonging to the Meloidogyne genus by taxonomic keys for the identification of parasitic plant nematodes (Zuckerman et al., 1990).
Identification of the nematode
The genomic DNA was isolated from 5000 nematodes (J2) using lysis with detergents followed by an extraction with phenol-chloroform-amyl alcohol (Sambrook and Russell, 2001). The specimens were collected through centrifugation at 16,000 g for 5 minutes and the pellet was resuspended in 200 µL of lysis buffer TSNT (2 % Triton X-100, 1% SDS, 100 mM NaCl, 10 mM, TrisHCl and 1mM EDTA, pH 8), 500 µL of saturated phenol with Tris-HCl and 100 µL of a mixture of chloroform: amyl alcohol (24:1). The suspension was mixed in vortex for 5 minutes and 200 µL of buffer TE 1X were added (10mM Tris-HCl and 1 mM EDTA, pH 8.0); the phases were separated by centrifugation at 16,000 g for 10 minutes, the DNA settled on the aqueous phase with pure ethanol. The DNA pellets were dried at room temperature for 10 minutes and were resuspended in 48 µL of TE buffer, 2 µL of 2 µg µL-1 RNase and quantified through agarose gel densitometry dyed with GelRed 1X (Biotium, Hayward, CA) solution, using the EDAS 290 system and the Kodak Digital Science 1D (Eastman Kodak Company, Rochester, New York) program.
Afragment of rDNAwas amplified by PCR using the ITS1 (5´-TGAACCGGGCAAAAGTCG-3´) initiator and the ITS2 (5´-TTAGTTTCTTTTCCTCCGCT-3´) initiator, directed at the regions 3´ and 5´ of the rDNA 18S and 28S, respectively (Stock et al., 2001; Meza-García et al., 2014). PCR was carried out in a TC020-24 MultiGene Mini (Labnet International, Inc., Edison, NJ) thermal cycler, with a final volume of 25 µL containing 1X buffer GoTaq PC (Promega, Madison, WI), 200 ng of genomic DNA, 200 µM of each dNTPs, 0.5 µM of each initiator, sterilized water and 1 U of GoTaq DNA Polymerase (Promega, Madison, WI). An amplification program of 32 cycles was used: 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 75 s, with a first denaturing stage of 94 °C for 3 min and a final elongation stage of 72 °C for 5 min. A plasmid that contains a fragment of ITS regions of Heterorhabditis indica Poinar, Karunakar & David 1992, (pGEMITSLZO) was used as a positive control of the PCR and the components of the PCR without DNA were used as negative control. The products of the PCR were analyzed in an electrophoresis with agarose gel 2.5 % dyed with GelRed 1X solution, using one unit of horizontal electrophoresis MSMUNIDUO (Sigma-Aldrich Co., St. Louis, MO), 100 volts for 60 minutes, and the product of the amplification was cloned in the pGEM(r)-T (Promega) vector, following the instructions of the manufacturer. The mixture of the ligation was used to transform cells of Escherichia coli DH5α, the transformed cells were selected through a white-blue test in agar slates LB with ampicillin (100µg mL-1), 100 µL IPTG (isopropil-β-D-1-thiogalactopyranoside) (2 mg mL-1). The resulting cell colonies of the transformed cells were analyzed in order to detect the presence of plasmid DNA through the extraction by alkaline lysis with an agarose gel 0.8 % dyed with GelRed 1X solution. Furthermore, the correct insertion of the amplified product in the pGEM(r)-T vector was confirmed through PCR using the ITS1 and ITS2 initiators, as was previously described.
The six positive plasmids by PCR of different colonies of E. coli were sequenced using the T7 and SP6 (Promega) initiator and a genetic analyzer ABI Prism 310 (Applied Biosystems, Foster City, CA) in the Unit of Molecular Biology of the Institute of Cellular Physiology of the Autonomous University of Mexico (UNAM).
The T7 and SP6 sequences (12 sequences in total) were aligned using the Contig Assembly Program module of the BioEdit v7.0.8.0 (Hall, 1999) program and the consensus sequence was compared to the nucleotide sequences in the databases using the nucleotide BLAST (BLASTn) tool of the National Center of Information about Biotechnology (NCBI). The genus and species of the organism with the nucleotide sequence most related to the consensus sequence was considered as the genus and species of the analyzed nematode.
Results and discussion
The DNA extraction method produced 25 µg of high quality genomic DNA (DO260/DO280 1.9) without any visible degradation. The analysis in agarose gel for the amplified product by PCR of genomic DNA showed a band of 608 pb, and for the positive control (H. indica) a band of 890 pb was obtained. Likewise, the analysis of PCR of 6 different plasmids of different transformed E. coli colonies confirmed the presence of the rDNA fragment inside the extracted plasmids (Figure 1B).
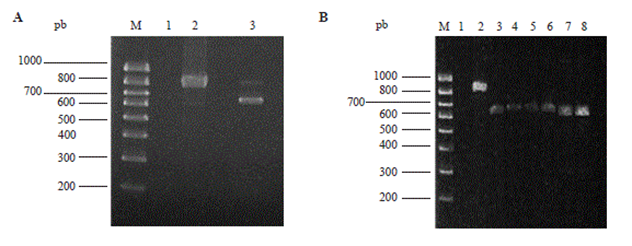
Figure 1 Amplification products by PCR from a genomic DNA of juvenile nematodes (A) or several plasmids of different transformed E. coli colonies (B). In A: Lane M, molecular sized marker Hyperlader IV (Bioline, London, UK); lane 1, negative control; lane 2, positive control (pGEMITSLZO); lane 3, the amplified product of 608 pb. In B: Lane M, molecular sized marker Hyperlader IV (Bioline, London, UL); lane 1, negative control; lane 2, positive control (pGEMITSLZO); lanes 3-8, plasmids of different transformants (608 pb).
The 66 identity percentages (BioEdit identity matrix) in pairs of the 12 nucleotide sequences (T7 and SP6 sequences of the 6 sequenced plasmids) oscillated between 99.6 and 100%, which allowed the calculation of a consensus sequence. Furthermore, the consensus nucleotide sequence presented an identity of 99.8% (2 differences, one generated by the ITS1 oligonucleotide) with a rDNA region of M. incognita (Figure 2) that includes the region 3´ of 18S, the internal transcribed spaces, the region 5.8S, the internal transcribed spacer 2 and a portion of the region and 5´ 28S. Therefore, the unknown nematode was identified as Meloidogyne incognita, confirming the presence of this species in the state of Nuevo León (México).

Figure 2 BLASTn Analysis of the consensus sequence of a fragment of rDNA of juvenile nematodes amplified through PCR (Query), that shows the alignment with a rDNA sequence of M. incognita (Sbjct), GenBank code AY438556.
In México, the Secretary of Agriculture, Livestock, Rural Developments, Fisheries and Food (SAGARPA) reported the presence of the nematode M. incognita, which is a significant pest for the state of Nuevo León (SENASICA-CNRF-Department of Plague Risk Analysis 2012). Usually, the species of the nematode of the Meloidogyne genus are identified by taxonomic keys in accordance to the morphological characteristics observed under the microscope (Powers, 2004). The morphological criteria represent a problem due to the variability and the need of experienced personnel in order to achieve the reliable identification of the species (Williamson et al., 1997). The identification of Meloidogyne spp. is based on the morphology of the adult females (Eisenback et al., 1981). However, a quick method for the identification of nematodes in the J2 stage could improve decision making for the optimal handling of infected crops (Powers and Harris, 1993).
In this work, the identification of a species of phytoparasitic root nematode using the ITS regions sequence of rDNA was achieved. The protocol used facilitates the identification or confirmation of the species, preventing the need for experienced personnel for the correct identification of the nematodes through the use of taxonomic keys.
Other DNA extraction methods used for the genomic DNA starting from the phytoparasitic nematodes, include the rupture of the cellular tissue through pressure (Powers and Harris, 1993), the use of lysis solutions with proteinase K (Williamson et al., 1997), and the extraction of DNA with phenol-chloroform followed by precipitation with ethanol (Williamson et al., 1997) or isopropanol (Blok et al., 1997). In this work, TSNT buffer was used as a lysis solution and the phenol-chloroform amyl alcohol was used for the extraction of DNA. This method allowed for the procurement of high quality genomic DNA for the amplification through PCR of the ITS regions of the rDNA.
The CDFA (California Department of Food and Agriculture, U.S.A.) includes the use of molecular methods in its protocol for the identification of the genus and species of different parasite nematodes present in plants, such as the Restriction Fragment Length Polymorphism of Amplified products (PCR-RFLP) (Dong, 2007). 5238 sequences of nematode ITS sequence regions are currently in the database of GenBank; among these sequences, 139 correspond to nematodes of the Meloidogyne genus. Therefore, the sequencing of ITS regions of problematic nematodes and the search in the nucleotide database is an appropriate alternative for their identification.
Conclusions
The molecular and bioinformatic methods are useful and quick tools for the diagnosis of phytoparasites of the Meloidogyne genus. This study provides a quick solution for the identification of nematodes in juvenile stages and also demonstrates that the amplification and analysis of ITS regions represents another option besides that of traditional methods. The clarity of the results allows for the identification of more species of nematodes, it is also an easy method, appropriate for both male and female juveniles, non-dependent of subjective criteria and does not require the immediate support of phytoparasitic nematode taxonomy experts.
Acknowledgements
We appreciate the support of this project with the key number CT2909-10 (PAICYT) of the Autonomous University of Nuevo León. J.J.-P., J.A.F.-G. y J.L.M.-G., and to CONACYT for their scholarships. We also thank MC Nabor González Garza for the supply and morphological identification of the biological material and to Sergio M. Salcedo Martínez, PhD. for his stylistic suggestions during the preparation of the manuscript.
REFERENCES
Adam MAM, Phillips MS, Blok VC. 2007. Molecular diagnostic key for identification of single juveniles of seven common and economically important species of root-knot nematode (Meloidogyne spp.). Plant Pathology. 56:190-197. [ Links ]
Agrios GN, 2005. Plant Pathology. Fifth Edition. Academic Press. New York, USA. 922p. [ Links ]
Blok VC, Philips MS, and Fargette M. 1997. Comparison of sequences from the ribosomal DNA intergenic region of Meloidogyne mayaguensis and other major tropical root-knot nematodes. Journal of Nematology. 29:16-22. [ Links ]
Cenis JL. 1993. Identification of four Major Meloidogyne spp. by Random Amplified Polymorphic DNA (RAPD-PCR). Phytopathology. 83:76-80. [ Links ]
Dong K. 2007. PCR-RFLP Identification of Globodera pallida, G. rostochiensis, Heterodera spp. and Meloidogyne spp. Available at: Available at: http://www.cdfa.ca.gov/plant/PPD/nematode_molecular.html . (Consulta, septiembre 2013). [ Links ]
Eisenback JD, Hirschmann H, Sasser JN, and Triantaphyllou AC. 1981. A guide to the four most common species of root-knot nematodes (Meloidogyne species), with a pictorial key. Journal of Nematology 13: 513-521. [ Links ]
Hall TA. 1999. BioEdit: A user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95-98. [ Links ]
Meza-García JL, Eías-Santos M, Cortez-Mondaca E, Guerrero-Olazarán M, Viader-Salvadó JM, Luna-Olvera HA, Maldonado-Blanco MG, Quintero-Zapata I, and Pereyra-Alférez B. 2014. Evaluation of Heterorhabditis indica (Rhabditida: Heterorhabditidae) Nematode Strain from Sinaloa, Mexico, against Bemisia tabaci Immatures under Laboratory Conditions. Southwestern Entomologist. 39(4), en impresión. [ Links ]
Orui Y. 1998. Identification of Japanese species of the genus Meloidogyne (Nematoda: Meloidogynidae) by PCR-RFLP analysis. Journal of Applied Entomology. 33: 43-51. [ Links ]
Powers TO, Harris TS. 1993. A polymerase chain reaction method for identification of five major Meloidogyne species. Journal of Nematology 25:1-6. [ Links ]
Powers TO. 2004. Nematode molecular diagnostics: From Bands to Barcodes. The Annual Review of Phytopathology. 42:367-383. [ Links ]
Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, Third Edition. Cold Spring Harbor Laboratory Press. New York, USA. 2100p. [ Links ]
SENASICA-CNRF-Departamento de Análisis de Riesgo de Plagas. 2012. Ficha técnica de Meloidogyne incognita. Available at: Available at: http://www.encuentra.gob.mx/resultsAPF.html?q=Meloidogyne%20incognita&client=sagarpa&ts=all&geo=0 . (Consulta, septiembre 2013). [ Links ]
Stock SP, Campbell JF, and Nadler SA. 2001. Phylogney of Steinernema travassos, 1927 (Cephalobina: Steinernema) inferred from ribosomal DNA sequences and morphological characters. Journal of Parasitology. 87:877-889. [ Links ]
Vovlas N, Mifsud D, Landa BB and Castillo P. 2005. Pathoge- nicity of the root-knot nematode Meloidogyne javanica on potato. Plant Pathology.54:657-664. [ Links ]
Williamson VM, Caswell-Chen EP, and Westerdahl FF. 1997. A PCR assay to identify and distinguish single juveniles of Meloidogyne hapla and M. chitwoodiJournal of Nematology. 29:9-15. [ Links ]
Wishart J, Phillips MS, and Blok VC.2002. Ribosomal Intergenic Spacer: A Polymerase Chain Reaction Diagnostic for Meloidogyne chitwoodi, M. fallax, and M. hapla. Phytopathology 92: 884-892. [ Links ]
Zijlstra C, Lever AEM, Uenk BJ and Silfhout CH. 1995. Differences between ITS Regions of Isolates of Root-knot Nematodes Meloidogyne hapla and M. chitwoodi. Phytopathology. 85: 1231-1237. [ Links ]
Zuckerman BM, Mai WF, and Krusberg LR. 1990. Plant Nematology Laboratory Manual Revised Edition 1990. University of Massachusetts Agricultural Experiment Station, Massachusetts. USA. 252 p. [ Links ]
Received: May 30, 2014; Accepted: December 15, 2014











 texto em
texto em 


