Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de fitopatología
versión On-line ISSN 2007-8080versión impresa ISSN 0185-3309
Rev. mex. fitopatol vol.26 no.1 Texcoco ene. 2008
Artículos científicos
Molecular and Biological Properties of a Putative Partitivirus from Jalapeño Pepper (Capsicum annuum L.)
Propiedades biológicas y moleculares de un posible partitivirus de chile jalapeño (Capsicum annuum L.)
Rodrigo A. Valverde and Dina L. Gutierrez
Louisiana State University Agricultural Center, Department of Plant Pathology and Crop Physiology, Baton Rouge, LA 70803 USA. GenBank Accession numbers for nucleotide sequence: DQ361008. Correspondence to: ravalve@lsu.edu
Received: September 17, 2007
Accepted: February 14, 2008
Abstract
Screening 50 greenhouse grown Jalapeño M (JM) (Capsicum annuum) pepper plants for the presence of pepper cryptic virus – 1 (PCV–1) by dsRNA extraction and analysis by electrophoresis, resulted in two JM lines. One was PCV–1 free and designated JM–N. The other one was PCV–1 positive and designated JM–P. Both lines were susceptible to natural infection by Cucumber mosaic virus and its satellite RNA. PCV–1 was not graft transmitted but it was transmitted both paternally (pollen) and maternally (ovule). Partial sequence of the RNA–dependent RNA polymerase gene was obtained from three overlapping cDNA clones derived from PCV–1 dsRNA. A search for aminoacid sequence similarity using the NCBI BLAST resulted in significant alignments with the RNA dependent RNA polymerase of Beet cryptic virus –3 and several putative members of the family Partitiviridae. Data presented indicate that PCV–1 is a distinct species of the family Partitiviridae.
Key Words: Cryptic virus, Cucumber mosaic virus, Beet cryptic virus–3, pepper cryptic virus–1, double–stranded RNA.
Resumen
Cincuenta plantas de chile Jalapeño M (JM) (Capsicum annuum) mantenidas en invernadero fueron evaluadas para la presencia del virus críptico: Pepper criptic virus–1 (PCV–1) a través del análisis del RNA viral de doble cadena, el cual dio como resultado dos líneas de JM. Una de ellas se designó JM–N debido a la ausencia del PCV–1 en esta línea. Mientras que la otra línea se designó JM–P, positiva a la presencia del PCV–1. Ambas líneas fueron susceptibles a la infección natural por el virus Cucumber mosaic virus y su RNA satelital. PCV–1 no se pudo transmitir por injerto, pero si por semilla, paternal (polen) y maternalmente (óvulo). La secuencia parcial del gen que codifica para la RNA polimerasa RNA–dependiente se obtuvo de tres clones superpuestos del cDNA proveniente del RNA viral de doble cadena del PCV–1. La similitud de la secuencia de aminoácidos se analizó a través del programa NCBI BLAST dando como resultado alineamientos significativos con la RNA polimerasa RNA–dependiente del virus Beet cryptic virus–3 y varios posibles miembros de la familia Partitiviridae. Los resultados de la presente investigación indican que PCV–1 es una especia distinta de la familia Partitiviridae.
Palabras Clave: Virus críptico, virus del mosaico del pepino, virus críptico–3 de la remolacha, virus críptico–1 del chile, RNA de doble cadena.
Healthy plants do not normally contain double–stranded RNA (dsRNA). In virus–infected plants, dsRNAs are found in the form of genomic segments of dsRNA viruses and replicative forms of single–stranded RNA viruses. DsRNAs have been used as diagnostic markers of viral infection in plants (Dodds et al., 1984; Valverde et al., 1990). The viral family Partitiviridae and the genus Endornavirus contain members that infect plants, have a dsRNA genome, are transmitted vertically through seed, but do not cause symptoms (Ghabrial et al., 2005; Gibbs et al., 2005). Endornaviruses have been reported in several plant species including rice (Oryza sativa L.), broad bean (Vicia faba L.), common bean (Phaseolus vulgaris L.), melon (Cucumis melo L.), and bell pepper (Capsicum annuum L.) (Fukuhara et al., 2006; Gibbs et al., 2005; Valverde and Gutierrez, 2007). The Partitiviridae is a family of isometric viruses with a segmented dsRNA genome (Ghabrial et al., 2005). Two genera: Alphacrytovirus and Betacryptovirus contain members that occur in plants and are transmitted through seed at high levels but cannot be transmitted by grafting or mechanical inoculation. Several cryptic viruses have been reported in different crop plants, but do not cause visible symptoms (Boccardo et al., 1987; Ghabrial et al., 2005). Alphacrytoviruses include those with two dsRNA segments of about 1.7 – 2.0 Kb and include: Alfalfa cryptic virus 1, White clover cryptic virus 1, and Beet cryptic virus 3 (BCV–3) (Boccardo et al., 1987; Ghabrial et al., 2005; Xie et al., 1993). Betacryptoviruses include those with two dsRNA segments of about 2.1 – 2.2 kbp with White clover cryptic virus 2 being the type member (Ghabrial et al., 2005). Some properties of a putative partitivirus found in Jalapeño M pepper designated pepper cryptic virus 1 (PCV–1) were reported previously (Arancibia and Valverde, 1995). The virus consists of spherical particles (30 nm), a coat protein of about 39 kDa, and two dsRNAs of 1.5 and 2.2 kbp (Arancibia and Valverde, 1995; Valverde and Gutierrez, 2007). In this paper we report partial sequence information, the relationship with some members of the Partitiviridae and some biological properties of PCV–1.
MATERIALS AND METHODS
Jalapeño M pepper (Petoseed, Woodland, CA, USA) (JM) plants, previously reported to contain a putative partitivirus (PCV–1) (Arancibia and Valverde, 1995) were grown under standard greenhouse conditions and used as source of dsRNA for transmission and cloning experiments. After screening 50 greenhouse grown JM plants for the presence of PCV–1 by dsRNA extraction and polyacrylamide gel electrophoresis (PAGE) analysis (Valverde et al., 1990), two types of plants were obtained. One type (two plants) was dsRNA free and a selected plant designated JM–N. The other type (48 plants) consisted of dsRNA–positive plants and a selected plant designated JM–P. Self pollination of the JM–N plant resulted in a progeny free of PCV–1 dsRNA (JM–N line), while self pollination of the JM–P plant yielded a progeny 100% with PCV–1 dsRNA (JM–P line). Simultaneous greenhouse evaluations of these two lines were conducted during different stages of plant growth (6, 8, 10, and 12 weeks after planting). Evaluations included plant growth habit, fruit color, shape, and fruit yield.
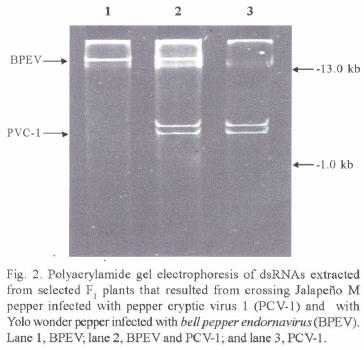
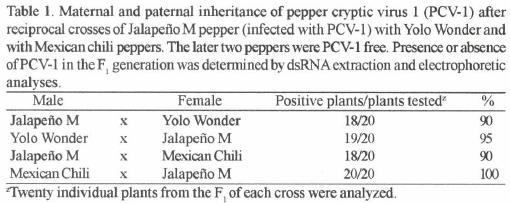
Transmission experiments. Graft transmission experiments to JM–N scions were conducted using JM–P rootstocks infected with Louisiana isolates (from pepper) of Tobacco etch virus (TEV) or Cucumber mosaic virus (CMV). TEV and CMV were used as transmission controls and to test their ability to assist the systemic spread of PCV–1. Four JM–P rootstocks infected with TEV and four infected with CMV were wedge–grafted with JM–N scions. Similarly, four other non–inoculated JM–P plants were grafted with JM–N scions. One month after grafting, symptoms were evaluated and scions tested for the presence of PCV–1 by electrophoretic analysis of dsRNA in polyacrylamide gels. Mexican Chili (MC) and Yolo Wonder (YW), two c. annuum cultivars that belong to two different horticultural types (ancho and bell, respectively) (Smith et al., 1987), were used in pollen and ovule transmission experiments. In previous studies, it was shown that YW was infected with an endornavirus (Valverde and Gutierrez, 2007) while MC tested free of dsRNA viruses. Reciprocal crosses between JM–P and YW, and JM–P and MC were conducted. At least four selected flowers from individual plants (maternal parent) were pollinated and tagged. Twenty plants (five from each of four successful crosses) from the F1 progeny of each cross were tested for the presence of PCV–1 by dsRNA extraction and PAGE analysis.
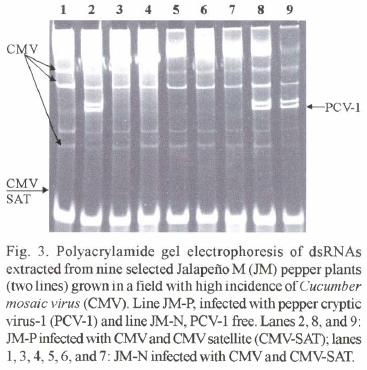
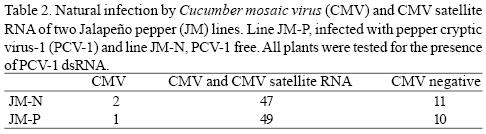
Natural infection of JM–N and JM–P by CMV. The susceptibility to CMV of field grown JM plants was evaluated. Seedlings from each JM line were grown in the greenhouse and tested for the presence or absence of PCV–1 by dsRNA analysis. Sixty (tested) seedlings of each line were transplanted in four rows (15 plants per row) at the Louisiana State University Agricultural Center experimental farm in Baton Rouge. All eight rows were randomly placed in a plot next to experimental plots of pepper breeding lines with high incidence (50 %) of CMV as determined by the Enzyme–linked immunosorbent assay (ELISA) using CMV antiserum. Two months later, foliar samples were collected from all the JM plants and tested for CMV infection using dsRNA analysis (Valverde et al., 1990).
DsRNA purification and cloning. DsRNA was extracted from JM–P plants, treated with RNAse A, DNAse I, and gel purified (from agarose gels) using the MinElute gel extraction kit (Qiagen, Valencia, CA, USA). DsRNA was denatured with dimethyl sulfoxide and used as template to generate a cDNA library using random hexadeoxynucleotide primers. cDNA was ligated into pGEM–T Easy Vector (Promega, Madison, WI, USA) and used to transform JM 109 competent cells. Several cDNA clones were obtained and selected clones were sequenced. The nucleotide sequences were determined by automated sequence analysis at Research Technology Support Facility of Michigan State University, (East Lansing, MI, USA) using a Perkin Elmer/Applied Biosystems 3100 capillary sequencer (Perkin Elmer, Foster City, CA). Derived amino acid sequences of three overlapping clones were compared with sequences in databases using the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1997). Multiple sequence alignment were conducted using the clustal series of programs available at the European Bioinformatics Institute Website (http://www.ebi.ac.uk) (Chenna et al., 2003).
RESULTS
Partial results of the dsRNA PCV–1 screening of 50 seedlings of JM are shown in Figure 1. The phenotype of JM–N was indistinguishable from JM–P. Evaluations of plant growth habit and fruit shape, size, and color did not result in differences. Fruit yield (number and weight per plant) did not show significant differences in both lines.

Transmission experiments. Graft transmission experiments were successful for CMV and TEV. Typical symptoms caused by these viruses on pepper were observed on all the JM–N scions one month after grafting. However. dsRNA analyzes of these scions, did not yield PCV–1 dsRNA. Electrophoretic analyses of dsRNAs clearly identified the presence or absence of PCV–1 dsRNA or YW dsRNA in the F1, plants that resulted from pollen and ovule transmission experiments (Fig. 2). Results of transmission experiments are shown in Table 1. PC V–1 dsRNA was transmitted both paternally and maternally. Slightly higher levels of transmission were obtained through the ovule than through pollen.
Natural infection by CMV. Natural infections by CMV and CMV satellite RNA were clearly and consistently detected by PAGE analyses of extracted dsRNA (Fig. 3). After two months, approximately 80% of the plants of both lines were infected with CMV and CMV satellite RNA (Table 2). Symptoms caused by CMV or CMV satellite RNA in both lines were undistinguishable. The symptoms consisted of mild foliar mosaic. Phenotypical differences between the tw o lines were not observed under field conditions.
Clones and sequence analyses. Three overlapping cDNA clones, J–30 (481 bp), J–64 (160 bp), and J–84 (783 bp) were sequenced. The region included in these clones (1069 bp) (GenBank accession No. DQ361008) contained an open reading frame predicted to encode a protein of 311 amino acids. A BLASTP search of the Uniprot database using derived amino acid sequences from this sequence resulted in significant similarity with predicted RNA–dependent RNA polymerase (RdRp) encoded by a plant virus member of the genus Alphacryptavirusand four putative plant partitiviruses. The highest scores were obtained with the RdRp encoded by a putative partitivirus from Japanese pear (Pyrus pyrifolia virus) (PPV, Q9ZWP7, 58% identity) (Osaki et al., 1998) followed by the alphacryptovirus Beet cryptic virus 3 (BC V–3, Q86632,54% identity) (Xie et al., 1993), and three putative partitiviruses: Pinus sylvestris cryptic virus (PSC V Q3ZDC6. 45% identity) (Veliceasa et al., 2006), Fragaria chiloensis cryptic virus (FCCV, Q4FCM7, 38% identity) (Tzanetakis and Martin, 2005) and Raphanus sativus cryptic virus 2 (RSCV–2, Q38L14, 37% identity) (Chen et al, 2006). Lower percentages were obtained with RdRp of fungal viruses (Fam. Partitiviridae, genus Partitivirus) including: Penicillium stoloniferum virus F (Q4G3H2, 26% identity), Mycovirus FusoV (Q83328, 26% identity), Discula destructive virus 2 (Q91HI8, 26% identity) and Gremmeniella abietina RNA virus MS2 (Q6GYB9, 24% identity). A Phylogram derived from the multiple alignments is shown in Figure 4. A 292 amino acid sequence of the putative RdRp region of PCV–1 was aligned with corresponding sequences of BCV–3 and three putative plant partitiviruses (Fig. 5). Five conserved motifs were identified.

DISCUSSION
Plant partitiviruses are transmitted only by seed (ovule and pollen). They are not graft transmissible because they are unable to move from cell to cell (Boccardo et al., 1987; Ghabrial et al., 2005). Biological and molecular properties of PCV–1 revealed similarities with plant partitiviruses. Attempts have been made to transmit plant partitiviruses from infected cultivars, however, in most cases different cultivars were targeted for transmission. Past attempts to transmit plant partitiviruses by graft inoculations did not test the possibility of a "helper effect" using conventional plant viruses (Boccardo et al., 1987). Results of graft inoculation experiments suggest that PCV–1 is not graft transmissible to a virus free line of the same pepper cultivar and that CMV or TEV infections do not assist the transmission. Pollen and ovule transmission studies conducted here confirmed a preliminary report on the transmission of PCV–1 (Arancibia and Valverde, 1995). Transmission rates were similar to those obtained with endornaviruses of rice and pepper (Horiuchi et al, 2003; Valverde and Gutierrez, 2007) which like the partitiviruses, accumulate in the cytoplasm. Although, transmission rates of PCV–1 through pollen (90%) were higher than the 30% rate reported by Arancibia and Valverde (1995). The reason for this discrepancy could be due to the fact that earlier transmission studies were conducted by crossing JM–P plants with pepper plants belonging to horticultural pepper types different from the ones used in this investigation. Natural infections of JM pepper by CMV and CMV satellite RNA did not appear to be affected by the absence or presence of PCV–1. Although not quantified, dsRNA levels of both, CMV and CMV satellite did not appear to be affected by the presence of PCV–1. Similarly, the presence of CMV and CMV satellite did not seem to affect the dsRNA yields of PCV–1. Differences in host reaction between the two lines were not observed with natural infections with CMV as in previous studies with mechanical inoculations with conventional viruses (Arancibia and Valverde, 1995). Multiple alignments of the conserved RdRp of PCV–1 and four putative partitiviruses (PPV, FCCV, PSCV, and RSCV–2), one alphacryptovirus (BCV–3) and four fungal partitiviruses, resulted in a higher percentage of similarity to plant partitiviruses than to fungal partitiviruses. This is illustrated with the unrooted phylogram (Fig. 4). However, it has been reported that two plant partitiviruses (White clover cryptic virus 1 and RSCV–2) have higher similarity to fungal viruses (Boccardo and Candresse, 2005; Chen et al., 2006). This supports the suggestion that the taxonomic status of the plant partitiviruses needs to be reevaluated (Veliceasa et al., 2006). The availability of more sequence information and biological properties of these viruses will help to clarify the taxonomic status of several putative partitiviruses. It has been shown that hypoviruses and endornaviruses can affect the physiology of the host (Gibbs et al, 2005; Nuss et al, 2005). Recently, Marquez et al. (2007) reported that a mutualistic interaction between a fungal endophyte (Curvularia protuberata Nelson and Hodges) and a tropical panic grass [Dichanhtelium lanuginosum (Elliott) Gould] allows both organisms to grow at high temperatures. Curvularia thermal tolerance virus, a putative Partitivirus infecting C. protuberata played a key role in conferring the heat tolerance to D. lanuginosum. Whether plant partitiviruses (alphacryptoviruses and betacryptoviruses) can provide beneficial adaptative traits such as pathogen tolerance or tolerance to extreme environments remains to be investigated. Comparative studies on the effect of plant partitiviruses in plants have not been possible due to the inability to infect plants with these viruses and the lack of plant lines of the same cultivar free of the virus. Although we obtained a virus–free line of JM (JM–N) and conducted limited experiments and preliminary observations, we have failed to demonstrate an effect. Further comparative and quantitative studies with these two lines need to be conducted.
ACKNOWLEDGMENTS
The authors wish to dedicate this article to the memory of Dr. Francis Can who conducted the field experiments.
LITERATURE CUED
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipmann, D.J. 1997. Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucleic Acids Research 25:3389–3402. [ Links ]
Arancibia, R.A., and Valverde, R.A. 1995 A cryptic virus from pepper. Plant Pathology 44:164–168. [ Links ]
Boccardo, G., and Candresse, T. 2005. Complete sequence of the RNA2 of an isolate of White clover cryptic virus 1, type species of the genus Alphacryptovirus. Archives of Virology 150:403–405. [ Links ]
Boccardo, G, Lisa, V, Luisoni, E., and Milne, R.G. 1987. Cryptic plant viruses. Advances in Virus Research 32:171–214. [ Links ]
Chen, L., Chen, J.S., Liu, L., Yu, X., Yu, S., Fu, T.Z., and Liu, W.H. 2006. Complete nucleotide sequences of three dsRNA segments from Raphanus sativus–root cv. Yipinghong. Archives of Virology 151:2077–2083. [ Links ]
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., and Thompson, J.D. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31:3497–3500. [ Links ]
Dodds, J.A., Morris, T.J., and Jordan, R.L. 1984. Plant viral double–stranded RNA. Annual Review of Phytopathology 22:151–168. [ Links ]
Fukuhara, T., Moga, R., Aoki, N., Yuki, C., Yamamoto, N., Oyama, N., Udagawa, T., Horiuchi, H., Miyazaki, S., Higashi, Y., Takeshita, M., Ikeda, K., Arakawa, M., Matsumoto, N., and Moriyama, H. 2006. The wide distribution of endornaviruses. Archives of Virology 151:995–1002. [ Links ]
Ghabrial, S.A., Buck, K.W., Hillman, B.I., and Milne, R.G 2005. Family Partitiviridae. pp. 581–590. In: C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, and L.A. Ball (eds.). Virus Taxonomy, VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press. London, UK. 1259 p. [ Links ]
Gibbs, M., Pfeiffer, P., and Fukuhara, T. 2005. Genus Endornavirus. pp. 603–605. In: C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, and L.A. Ball (eds.). Virus Taxonomy, VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press. London, UK. 1259 p. [ Links ]
Horiuchi, H., Moriyama, H., and Fukuhara, T. 2003. Inheritance of Oryza sativa endornavirus in F1 and F2 hybrids between japonica and indica rice. Genes and Genetic Systems 78:229–234. [ Links ]
Marquez, L.M., Redman, R., Rodríguez, R.J., and Roossinck, M.J. 2007. A virus in a fungus in a plant: three–way symbiosis required for thermal tolerance. Science 315:513–515. [ Links ]
Nuss, D.L., Hillman, B.I., Rigling, D., and Suzuki, N. 2005. Family Hypoviridae. pp. 597–601. In: C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, and L.A. Ball (eds.). Virus Taxonomy, VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press. London, UK. 1259 p. [ Links ]
Osaki, H., Kudo, A., and Ohtsu, Y. 1998. Nucleotide sequence of seed–and pollen–transmitted double–stranded RNA which encodes a putative RNA–dependent RNA polymerase, detected from Japanese pear. Bioscience, Biotechnology and Biochemistry 62:2101–2106. [ Links ]
Smith, P.G., Villalon, B., and Villa, P.L. 1987. Horticultural classification of peppers grown in the United States. HortScience 22:11–13. [ Links ]
Tzanetakis, I.E., and Martin, R.R. 2005. Fragaria chiloensis cryptic virus: A new strawberry virus found in Fragaria chiloensis from Chile. Plant Disease 89:1241. [ Links ]
Valverde, R.A., and Gutierrez, D.L. 2007. Transmission of a dsRNA in bell pepper and evidence that it consists of the genome of an endornavirus. Virus Genes 35:399–403. [ Links ]
Valverde, R.A., Nameth, S.T., and Jordan, R.L. 1990. Analysis of double–stranded RNA for plant virus diagnosis. Plant Disease 74:255–258. [ Links ]
Veliceasa, D., Enunlu, N., Kos, P.B., Koster, S., Beuther, E., Morgun, B., Deshmukh, S.D., and Lukacs, N. 2006. Searching for a new putative cryptic virus in Pinus sylvestris L. Virus Genes 32:177–186. [ Links ]
Xie, W.S., Antoniw, J.F., and White, R.F. 1993. Nucleotide sequence of beet cryptic virus 3 dsRNA2 which encodes a putative RNA–dependent RNA polymerase. Journal of General Virology 74:1467–1470. [ Links ]














