Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Similars in
SciELO
Similars in
SciELO
Share
Acta zoológica mexicana
On-line version ISSN 2448-8445Print version ISSN 0065-1737
Acta Zool. Mex n.86 Xalapa Aug. 2002
Article
Exploring the relation between genetic structure and habitat heterogeneity in the rodent Liomys pictus from Chamela, Jalisco
Ella Vázquez-Domínguez, Gerardo Ceballos and Daniel Piñero
Instituto de Ecología, UNAM. Apartado Postal 70-275, Ciudad Universitaria. México, D.F. 04510, MÉXICO. E-mail: evazquez@miranda.ecologia.unam.mx
Recibido: 3 de mayo 2001
Aceptado: 12 de octubre 2001
Resumen
Se estudiaron poblaciones locales del ratón espinoso, Liomys pictus, de la selva tropical caducifolia y subcaducifolia en Chamela, Jalisco, México, donde L. pictus presenta marcadas fluctuaciones en densidades poblacionales y altas tasas de intercambio poblacional. Dichas características están relacionadas directamente con la severa estacionalidad ambiental y la heterogeneidad característica de estas selvas. Con el objetivo de revisar la hipótesis que señala que el nivel de variación genética de una población puede estar correlacionado con el grado de heterogeneidad ambiental, se evaluaron las diferencias genéticas entre poblaciones de estos dos ambientes contrastantes (selvas caducifolia y subcaducifolia). Se analizaron 30 loci con electroforesis en gel de almidón. Las frecuencias alélicas, el número de loci heterócigos y los valores de heterocigosidad observada y esperada no fueron significativamente diferentes entre subpoblaciones. Las diferencias entre éstas y la dispersión de individuos también fueron evaluadas con una prueba de asignación basada en genotipos individuales. Setenta y dos por ciento de los individuos fueron asignados correctamente a su población de origen, lo que se interpretó como indicativo de una baja diferenciación interpoblacional. Los cambios en la variabilidad genética que se observan como respuesta a las fluctuaciones poblacionales y altas tasas de intercambio, aunado al movimiento de los animales (se encontró una marcada dispersión, significativamente mayor en machos), pudieron cancelar o atenuar las divergencias entre subpoblaciones. Se observó además cierta endogamia, lo cual puede interpretarse como efecto de Wahlund, que resultaría de mezclar subpoblaciones con frecuencias alélicas diferentes. Por otro lado, sugerimos que la filopatría de las hembras y la dispersión predominantemente de los machos, que representan componentes característicos de la estructura social en L. pictus, puede resultar ya sea en la dispersión de los individuos (estructuración poblacional) o en la mezcla de subpoblaciones (efecto de Wahlund). Este tipo de cambios poblacionales y genéticos son aspectos importantes en muchas poblaciones de pequeños mamíferos, por lo que deben tomarse en cuenta cuando se evalúan la demografía y estructura genética.
Palabras clave: Liomys pictus, roedores heterómidos, variabilidad genética, heterogeneidad ambiental, selva tropical seca, isoenzimas.
Abstract
Local populations of the spiny pocket mice, Liomys pictus, were sampled from the tropical deciduous and semideciduous forests from Chamela, Jalisco, Mexico, where L. pictus experiences profound population fluctuations and high rates of population turnover, associated with the strong environmental seasonality and heterogeneity characteristic of these forests. In order to review the hypothesis stating that the level of genetic variation in a population is correlated with the degree of environmental heterogeneity, the genetic differences between subpopulations from these two contrasting habitats were evaluated. Thirty presumptive gene loci were analyzed using starch-gel electrophoresis. Allele frequencies, number of heterozygous loci and observed and expected heterozygosity values were not statistically different between subpopulations. Population differences and sex-biased dispersal were also evaluated with an assignment test based on individual genotypes. Seventy-two percent of individuals were correctly assigned to their population of origin, which we considered indicative of low interpopulation differentiation. The short-term changes in genetic variability, as a response to the fluctuations in population density and high rates of population turnover, together with movement of individuals (a marked male-biased dispersal was observed), could conceal or buffer divergences between these subpopulations. L. pictus subpopulations were also characterized by inbreeding, which might be interpreted as a Wahlund effect, resulting from mixing of subpopulations of differing allelic frequencies. On the other hand, we suggested that female philopatry and male-biased dispersal, main components of the social structure of this species, may result in either dispersion of individuals (population structuring) or mixing subpopulations (Wahlund effect). Short-term population and genetic changes are important aspects of many small mammalian populations, which should be considered when assessing demography and genetic structure.
Key Words: Liomys pictus, heteromyid rodents, genetic variability, habitat heterogeneity, tropical dry forests, allozymes.
Introduction
The family Heteromyidae comprises the group of exclusively New World rodents that includes the kangaroo rats, pocket mice and kangaroo mice. The evolutionary diversification of this group in heterogeneous landscapes provides examples of genetic, ecological and biogeographic processes associated with their speciation and differentiation (Genoways & Brown 1993). Nevertheless, with important exceptions (Rogers 1990, Patton & Rogers 1993), we lack detailed information regarding their genetic structure, as well as about the relation of that genetic variability with factors such as environmental features, population structure and social organization.
The spiny pocket mice Liomys pictus, an endemic heteromyid species of western and southern Mexico (Genoways & Brown 1993), shows complex and specialized morphological, behavioral and physiological characteristics (Ceballos 1991, French 1993, Randall 1993, Mendoza 1997, Vázquez-Domínguez et al. 1998). It has been extensively studied in the dry deciduous and semideciduous forests of Chamela, Jalisco, Mexico (Rogers 1990, Ceballos 1991, Mendoza 1997, Vázquez-Domínguez 1997); however, there is scarce information about the genetic structure of this species regarding its populations in Chamela (Rogers 1990, Rogers & Engstrom 1992). These forests show a strong climatic seasonality determined by the amount and pattern of temporal distribution of rainfall, which has direct effects on phenology and food availability. They also reflect a marked gradient, from deciduous to semideciduous forest, of increasing spatial structural complexity and decreasing temporal seasonality, owing to the greater availability of water in the latter (Bullock 1986).
It has been shown that patterns of distribution and abundance of L. pictus are directly related to the environmental seasonality characteristic of these forests: it is the most abundant species in the deciduous forest where it is present throughout the dry season; density is higher at the end of the rainy season and beginning of the dry season; and reproduction and population fluctuations are strongly influenced by seasonality in food availability (Ceballos 1991, Mendoza 1997, Vázquez-Domínguez 1997). Because of its specialization, this species is able to maintain populations in the deciduous forest and cope with the high environmental seasonality and heterogeneity (French 1993, Mendoza 1997, Vázquez-Domínguez et al. 1998). The influence of habitat heterogeneity (i.e. climatic seasonality and habitat complexity) as an ecological and evolutionary force has long been recognized and it plays a key role in processes at population, community and ecosystem levels (Avise 1994). Also, habitat heterogeneity can affect, among other things, genetic polymorphisms (Patton & Feder 1981, Hedrick 1986). Accordingly, populations are composed of individuals living in mosaics of macro- and microhabitats that influence individual fitness and largely determine population dynamics (Avise 1994, Johannesson & Tatarenkov 1997).
Different studies with vertebrates have shown a positive correlation between environmental diversity and genetic polymorphisms (Schmitt et al. 1995, Johannesson & Tatarenkov 1997), although this relationship has not always been found (Ayala et al. 1975). Also, studies of intraspecific genetic variation over heterogeneous habitats suggest habitat-specific variation at local or even microscales (Nevo et al. 1994, Gebczynski & Ratkiewicz 1998). There are few examples of these kind of associations in vertebrates and even fewer in mammals (Patton & Feder 1981, Scribner & Chesser 1993, Schmitt et al. 1995). Similarly, it has been shown that the social organization and the concomitant dispersal and breeding patterns of a population, for mammals in particular, have important consequences for the apportionment of genetic variance and distribution of genotypic proportions within and among populations or lineages (Chesser 1991, Mathews & Porter 1993, Scribner et al. 1997). Accordingly, the structure of mammalian populations is influenced strongly by social organization and environmental constraints (Petit et al. 1997).
The purpose of the present work was to review the hypothesis concerning whether the level of genetic variation in a population is correlated with the degree of environmental heterogeneity (McDonald & Ayala 1974). On the other hand, considering the results observed in this study, we suggested some possible consequences of population structure and social organization on the genetic structure of the species.
Material and methods
Field work. Rodents were trapped at the Chamela-Cuixmala Biosphere Reserve, which is located in the southern portion of the State of Jalisco, on Mexico's west coast (19º25'N and 105º00'W). The vegetation is predominantly tropical deciduous forest (hereafter dry forest) and semideciduous forest (arroyo forest; Rzedowski 1978). The climate is characterized by its dry-wet seasonality, with an average monthly temperature of 24.9ºC and a mean precipitation of 748 mm/year, 80% of which occurs between July and October (Bullock 1986). The rest of the year is characterized by a marked dry season, when most plants shed their leaves.
Individuals of L. pictus were collected in the dry and arroyo forests, at two 0.8-ha grids per forest, for a total of 52 individuals per forest. Dry and arroyo trapping areas were approximately 5 km apart. Mice were trapped using Sherman live-traps, baited with a mixture of rolled oats, peanut butter and vanilla extract. Individuals were sexed and weighed to the nearest 0.1 g. Blood and tissue samples of all individuals were stored in liquid nitrogen; subsequently, tissue samples were homogenized in a buffered solution (pH 6.8) of Trizma base and EDTA and stored at -70ºC until electrophoresis was performed.
Genetic variability. Blood and tissue samples were used to assay 34 proteins with horizontal starch-gel electrophoresis. Good resolution was obtained for 19 proteins encoding 30 presumptive loci. Stains and buffers were used as described by Selander et al. (1971), Pasteur et al. (1988) and Teska et al. (1990). Buffer systems and enzymes studied are summarized in the Appendix. For accuracy, reference samples were run on each gel to ensure consistent scoring.
Genetic variability within local populations was calculated and expressed as the mean number of alleles per locus (A), the percentage of polymorphic loci per population (P, using the 95% criterion; Nei 1987), and the proportion of loci heterozygous per individual at each locality (H, direct count estimate; Hedrick 1983). These estimates were first calculated for each subpopulation and then for the entire sample. A G test was used to evaluate differences in observed and expected heterozygosity values between dry and arroyo forests (Wayne 1990).
To examine heterozygote proportions within and between dry and arroyo populations, FST, FIS and FIT estimates (Hedrick 1983) were calculated. To evaluate if these indices were different from zero, a X2 test was performed following Hedrick (1983) and Workman & Niswander (1970); and because with multiple comparisons the chance of an error increases, we used the Bonferroni's approximation (a/m, Fry 1993) to calculate a suitable significance level. Differences between dry and arroyo forests regarding the number of heterozygous loci of individuals were examined with a t-test (Sokal & Rohlf 1981). In order to avoid sex biased results, we tested if males and females showed different number of heterozygous loci with a G test for contingency tables (Wayne 1990).
Different applications of assignment tests have been successfully used as an approach to estimate population differentiation and sex-biased dispersal (Paetkau et al. 1995, Andersen et al. 1997, Waser & Strobeck 1998, Mossman & Waser 1999). Based on the variation of the 23 polymorphic loci, we used the assignment test of Paetkau et al. (1995), which determines how indicative an individual's genotype is of the population from which it was sampled (individual's assignment index). An individual is assigned to the population that has the highest likelihood of containing a member with the observed multilocus genotype (for details for computing the test see Paetkau et al. 1995).
We explored the movement of individuals between sites from a genetic point of view by two means: first, we estimated the number of migrants per generation (genetic flow Nm) through an indirect method proposed by Crow and Aoki (1984), which considers the number of subpopulations: FST = 1/(4aNm+1), where N is the population size, m is the fraction of N substituted by migrants and a = (n/n-1)2, where n is the number of subpopulations. Second, we carried out the assignment test of Paetkau et al. (1995) for the two populations together to detect a bias in dispersal between males and females, applying the modification proposed by Favre et al. (1997). The latter allows the identification of immigrants because individuals with a lower assignment index have rare genotypes and thus are potentially recent immigrants (Favre et al. 1997, Mossman & Wasser 1999).
Results
In the dry forest 29 males and 23 females were collected, and 25 and 27, respectively, in the arroyo forest. Average weight of males was 42.3 ± 5.6 g and 41.2 ± 6.3 g, and of females 38.0 ± 4.8 g and 38.5 ± 4.3 g, for dry and arroyo forests, respectively.
Twenty-three of 30 loci examined were polymorphic (Table 1), with a mean number of alleles per locus of 2.0 and 2.1, a percentage of polymorphic loci of 76.7 and 73.3, and an average observed heterozygosity value of 0.085 and 0.094, for dry and arroyo forests respectively. The estimates for the entire sample (A= 2.1, P= 73.3, H= 0.089) were similar and not statistically different to subpopulation estimates. Allele frequencies differed significantly between dry and arroyo forests for five loci (Aco1, G6pd1, G6pdD2, Ldh2, and Pepa2; Table 1) and the observed and expected heterozygosity values were not significantly different between the two forests (p> 0.05).
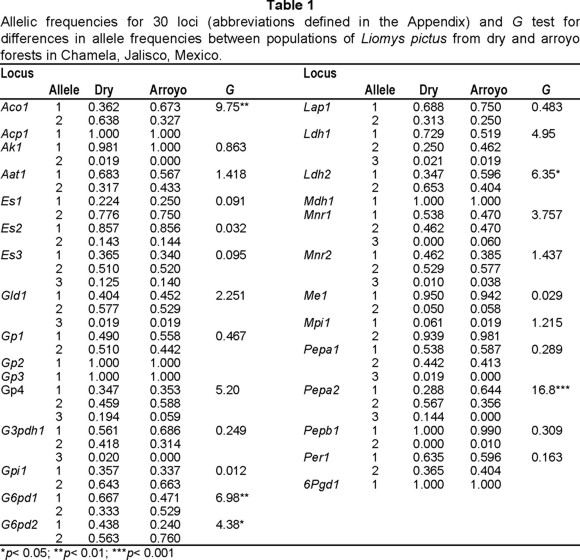
Mean FST = 0.023 was not significantly different from zero after Bonferroni's correction (p> 0.05), although some slight differentiation between subpopulations for heterozygote proportions was observed (values showed significant differentiation in 30% of the polymorphic loci: Aco1, Gp4, G6pd1, G6pd2, Ldh1, Ldh2 and Pepa2; p< 0.01). Mean FIS and FIT showed significant positive values (0.724 and 0.730, respectively; p< 0.001). Individual mice were heterozygous at as many as 8 loci and the number of heterozygous loci did not differ significantly between males and females, nor between dry and arroyo populations (p> 0.05).
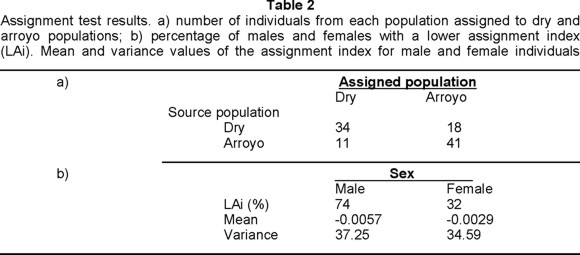
Assignment test results showed that a total of 75 individuals (72%) were assigned to the sampling area where they had been collected, with a higher number of misassigned individuals for the dry forest (Table 2). Regarding dispersal of individuals, Nm results showed that, from a genetic stand point, 2.6 individuals on average move between the two subpopulations per generation. Moreover, 74% percent of male individuals showed a lower assignment index compared to only 32% of the females, and male assignment indices also had a lower mean and higher variance (Table 2), all of which indicates a male-biased dispersal (Favre et al. 1997)
Discussion
Population numbers, species composition and demographic patterns that characterize rodent populations in Chamela are directly correlated to the fluctuating environment and seasonal patterns in resources availability present throughout the year (Ceballos 1991, Mendoza 1997, Vázquez-Domínguez 1997); such characteristics would agree with the genetic variability-heterogeneity hypothesis. Despite this, dry and arroyo populations in our study showed no significant differences for such genetic estimates as number of heterozygous loci per individual and observed and expected heterozygosity, and only slightly for allele frequencies.
The assignment test used here asks whether sufficient differences exist between subpopulations to make an individual's genotype characteristic, or even diagnostic, of the population from which it came (Patekau et al. 1995). Thus, it provides a measure of population differentiation by indicating how many individuals are assigned to the area in which they were sampled. Accordingly, in the present results 65% and 79% of the animals were correctly assigned to the dry and arroyo populations, respectively. In the present case we think that the high percentage of misassigned individuals will indicate low interpopulation differentiation, despite the fact that some studies with similar values had been reported as having highly structured populations (Patekau et al. 1995, Primmer et al. 1999). Although individual genetic descriptors like the assignment test have been used with allozymes (Andersen et al. 1997), their common use with highly variable loci (e.g. microsatellites) makes them remarkably reliable, and likely best suited than for allozyme data, thus, only values closer to the one hundred percent will confidently indicate spatial structuring of populations.
That no differences were found between subpopulations could be partially explained in terms of the spatial and temporal shifts in genetic variability expected in fluctuating populations with overlapping generations, in which short-term changes in genetic variability can be responses to fluctuations in population density and high rates of population turnover (Smith et al. 1975, Harrison and Hastings 1996). Such changes could conceal possible genetic differences in these L. pictus subpopulations that were sampled in a single point in time. Also, the estimates of gene flow (Nm) observed in the present study may buffer divergences during stages with lower population sizes (e.g. during the long draught months when population numbers are drastically lowered; Mendoza 1997, Vázquez-Domínguez 1997).
Our results also indicate that the subpopulations of L. pictus studied are characterized by inbreeding. One direct interpretation is that the observed inbreeding could be due to a Wahlund effect, resulting from pooling genotypic data by mixing of populations of differing allelic frequencies. The most plausible explanation for this would be that the populations sampled represent two cryptic species, a possibility that we cannot rule out with the present data.
These results could also be related to the characteristic population structure and social organization of this species. Different studies have demonstrated that the structure of mammalian populations is strongly influenced by social organization. They have also evidenced some consequences of population structure and social organization on genetic variation (Chesser 1991, Scribner et al. 1991, Mathews & Porter 1993, Petit et al. 1997, Scribner et al. 1997). This is, inbreeding is enhanced by spatial structuring, but can occur in its absence (Scribner et al. 1991) and, although commonly perceived as consanguineous matings, can result from matings among non familial relatives, related because of low effective population size (Chesser & Ryman 1986). Breeding among relatives also may occur in populations that show strong site fidelity (philopatry); in these, despite male movements and gene flow among sedentary female groups, inbreeding can occur if dominant males tend to sire most offspring produced within certain female groups or matriarchical lines (Chesser 1991, Scribner et al. 1991). High rates of gene flow among subpopulations may counter deleterious levels of inbreeding.
In this respect, female philopatry and male-biased dispersal have been shown for L. pictus in Chamela (Mendoza 1997). Also, reproductively active females live alone and maintain distinct home ranges, while male home ranges overlap those of other males and females (Randall 1993, Mendoza 1997). Our results confirm that animals move considerably between subpopulations and that this dispersal is predominantly male-biased. Hence, female philopatry and male-biased dispersal, together with the fluctuating variation in reproduction and population sizes observed in L. pictus in Chamela, may result in variation in juxtaposition of males and females during breeding periods (Scribner et al. 1997). This in turn could result in either dispersion of individuals of different subpopulations, with which some degree of spatial structuring would be observed, or mixing gene pools and subsequent Wahlund effect.
Differences in genetic characteristics are known to occur in vertebrates related to sexes, ages, body size, cohorts and population dynamics, in small mammals and even in relatively long-lived species (Massey & Joule 1981, Scribner & Chesser 1993). It also has been suggested that mammalian populations, due to their genetically heterogeneous dynamics, can form subpopulations with different genetic characteristics (Scribner et al. 1991). Accordingly, the genetic structure observed in Liomys pictus in the present study supports the importance of considering short-term populations structure and genetic changes in the study of the interactions among demography, genetic structure and behavior.
Acknowledgements
This study resulted from research conducted in partial fulfillment of requirements for the Ph.D. degree in the Instituto de Ecología, Universidad Nacional Autónoma de México by EV. We thank the staff of the Chamela-Cuixmala Biosphere Reserve for providing lodging, and laboratory and field work facilities. León Cázares and Olivia Reynoso for technical assistance with the homogenization methods and laboratory facilities. Two anonymous reviewers made helpful comments on the manuscript. Partial funding was provided by Idea Wild and by the Fundación Ecológica de Cuixmala, A.C. First author's scholarship was provided by the National Council of Science and Technology (CONACYT; registration number 86298).
Literature cited
Andersen, L.W., L. Holm, H.R. Siegismund, B. Clausen, C.C. Kinze & V. Loeschcke. 1997. A combined DNA-microsatellite and isozyme analysis of the population structure of the harbor porpoise in Danish waters and West Greenland. Heredity 78:270-276. [ Links ]
Avise, J.C. 1994. Molecular markers, natural history, and evolution. Chapman and Hall, New York. 511 pp. [ Links ]
Ayala, F.J., J.W. Valentine, D. Hedgecock & L.G. Barr. 1975. Deep-sea asteroids: high genetic variability in a stable environment. Evolution 29:203-212. [ Links ]
Bullock, S.H. 1986. Climate of Chamela, Jalisco, and trends in the south coastal region of Mexico. Arch. Met. Geogr. Biocl. Ser. B 36:297-316. [ Links ]
Ceballos, G. 1991. Comparative natural history of small mammals from tropical forests in western Mexico. J. Mamm. 71:263-266. [ Links ]
Chesser, R.K. 1991. Genetic diversity and female philopatry. Genetics 127:437-447. [ Links ]
Chesser, R.K. & N. Ryman. 1986. Inbreeding as a strategy in subdivided populations. Evolution 40:616-624. [ Links ]
Crow, J.F. & K. Aoki. 1984. Group selection for a polygenic behavioral trait: estimating the degree of populations subdivision. Proc. Natl. Acad. Sci. USA 81:6073-6077. [ Links ]
Favre, F., F. Balloux, J. Goudet & N. Perrin. 1997. Female-biased dispersal in the monogamous mammal Crocidura rassula: evidence from field data and microsatellite patterns. Proc. R. Soc. London Ser. B 264:127-132. [ Links ]
French, A.R. 1993. Physiological ecology of the Heteromyidae: economics of energy and water utilization. Pp. 509-538. In H.H. Genoways and J.H. Brown (eds). Biology of the Heteromyidae. Special Publication 10. The American Society of Mammalogists, USA. [ Links ]
Fry, J.C. (Ed.). 1993. Biological data analysis. A practical approach. Oxford University Press, Oxford. 418 pp. [ Links ]
Genoways, H.H. & J.H. Brown. (Eds.). 1993. Biology of the Heteromyidae. Special Publication 10. The American Society of Mammalogists, USA. 719 pp. [ Links ]
Gebczynski, M. & M. Ratkiewicz. 1998. Does biotope diversity promote an increase of genetic variation in the bank vole population? Acta Theriol. 43:163-173. [ Links ]
Harrison, S. & A. Hastings. 1996. Genetic and evolutionary consequences of metapopulation structure. Trends Ecol. Evol. 11:180-183. [ Links ]
Hedrick, P.W. 1983. Genetics of populations. Jones and Bartlett, Boston. 553 pp. [ Links ]
––––––––––. 1986. Genetic polymorphism in heterogeneous environments: A decade later. Annu. Rev. Ecol. Syst. 17:535-566. [ Links ]
Johannesson, K. & A. Tatarenkov. 1997. Allozyme variation in a snail (Littorina saxatilis) - Deconfounding the effects of microhabitat and gene flow. Evolution 51:402-409. [ Links ]
Massey, D.R. & J. Joule. 1981. Spatial-temporal changes in genetic composition of deer mouse populations. Pp. 180-201. In M.H. Smith and J. Joule (eds). Mammalian population genetics. University of Georgia Press, Athens. [ Links ]
Mathews, N.E. & W.F. Porter. 1993. Effect of social structure on genetic structure of free-ranging white-tailed deer in the Adirondack mountains. J. Mamm. 74:33-43. [ Links ]
McDonald, J.F. & F.J. Ayala. 1974. Genetic response to environmental heterogeneity. Nature 250:572-574. [ Links ]
Mendoza, D.A. 1997. Efecto de la adición de alimento en la dinámica de poblaciones y estructura de comunidades de pequeños mamíferos en un bosque tropical caducifolio. Tesis de Maestría. Facultad de Ciencias, Universidad Nacional Autónoma de México, México. 100 pp. [ Links ]
Mossman, C.A. & P.M. Waser. 1999. Genetic detection of sex-biased dispersal. Mol. Ecol. 8:1063-1067. [ Links ]
Nei, M. 1987. Molecular evolutionary genetics. Columbia Univ. Press, New York. 512 pp [ Links ]
Nevo, E., T. Krugman & A. Beiles. 1994. Edaphic natural selection of allozyme polymorphisms in Aegilops peregrina at a Galilee microsite in Israel. Heredity 72: 109-112. [ Links ]
Paetkau, D., W. Calvert, I. Stirling & C. Strobeck. 1995. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4:347-354. [ Links ]
Pasteur, N., G. Pasteur, F. Bonhomme, J. Catalan & J. Britton-Davidian. 1988. Practical isoenzyme genetics. John Wiley and Sons, New York. 215 pp [ Links ]
Patton, J.L. & J.H. Feder. 1981. Microspatial genetic heterogeneity in pocket gophers: non-random breeding and drift. Evolution 31:697-720. [ Links ]
Patton, J.L. & D.S. Rogers. 1993. Biochemical genetics. Pp. 259-269. In H.H. Genoways and J.H. Brown (eds). Biology of the Heteromyidae. Special Publication 10. The American Society of Mammalogists, USA. [ Links ]
Petit, E., S. Aulagneir, R. Bon, M. Dubois & B. Crouau-Roy. 1997. Genetic structure of populations of the mediterranean Mouflon (Ovis gmelini). J. Mamm. 78:459-467. [ Links ]
Primmer, C.R., T. Aho, J. Phronen, et al. 1999. Microsatellite analysis of hatchery stocks and natural populations of Arctic charr, Salvelinus alpinus, from the Nordic region: implications for conservation. Hereditas 130:277-289. [ Links ]
Randall, J.H. 1993. Behavioral adaptations of desert rodents (Heteromyidae). Animal Behav. 45:263-287. [ Links ]
Rogers, D.S. 1990. Genic evolution, historical biogeography, and systematic relationships among spiny pocket mice (Subfamily Heteromyinae). J. Mamm. 71:668-685. [ Links ]
Rogers, D.S. & M.D. Engstrom. 1992. Genic differentiation in spiny pocket mice of the Liomys pictus species-group (family Heteromyidae). Can. J. Zool. 70:1912-1919. [ Links ]
Rzedowski, J. 1978. Vegetación de México. Limusa, México. 432 pp. [ Links ]
Schmitt, L.H., D.J. Kitchener & R.A. How. 1995. A genetic perspective of mammalian variation and evolution in the Indonesian archipielago: Biogeographic correlates in the Fruit bat genus Cynopterus. Evolution 49:399-412. [ Links ]
Scribner, K.M. & R.K. Chesser. 1993. Environmental and demographic correlates of spatial and seasonal genetic structure in the eastern cottontail rabbit (Sylvilagus floridanus). J. Mamm. 74:1026-1044. [ Links ]
Scribner, K.M., M.H. Smith, R.A. Garrot & L.H. Carpenter. 1991. Temporal, spatial, and age specific changes in genotypic composition of mule deer. J. Mamm. 72:126-137. [ Links ]
Scribner, K.M., M.H. Smith & R.K. Chesser. 1997. Spatial and temporal variability of microgeographic genetic structure in white-tailed deer. J. Mamm. 78:744-755. [ Links ]
Selander, R.K., M.H. Smith, S.Y. Yang, W.J. Johnson & J.B. Gentry. 1971. Biochemical polymorphism and systematics in the genus Peromyscus. I. Variation in the oldfield mouse (Peromyscus polionotus). Studies in Genetics IV. Univ. Texas Publ. 7103:49-90. [ Links ]
Smith, M.H., C.T. Garten, Jr. & P.R. Ramsey. 1975. Genic heterozygosity and population dynamics in small mammals. Pp. 85-102. In C.L. Markert (ed). Isozymes IV. Genetics and Evolution. Academic Press, New York. [ Links ]
Sokal, R.R. & F.J. Rohlf. 1981. Biometry: the principles and practice of statistics in biological research. Second ed. W.H. Freeman and Company, San Francisco. 859 pp. [ Links ]
Teska, W.R., M.H. Smith & J.M. Novak. 1990. Food quality, heterozygosity, and fitness correlates in Peromyscus polionotus. Evolution 44:1318-1325. [ Links ]
Vázquez-Domínguez, E. 1997. Patrones de heterocigosidad y su relación con componentes de adecuación del roedor Liomys pictus en Chamela, Jalisco. Tesis de Doctorado. Instituto de Ecología, UNAM. México. 164 pp. [ Links ]
Vázquez-Domínguez, E., D. Piñero & G. Ceballos. 1998. Heterozygosity patterning and its relation to fitness components in experimental populations of Liomys pictus from tropical forests in western Mexico. Biol. J. Linn. Soc. 65:501-514. [ Links ]
Waser, P.M. & C. Strobeck. 1998. Genetic signatures of interpopulation dispersal. Trends Ecol. Evol. 13:43-44. [ Links ]
Wayne, W.D. 1990. Applied nonparametric statistic. Second ed. PWS-KENT Publishing Company, Boston. 635 pp. [ Links ]
Workman, P.L. & J.D. Niswander. 1970. Population studies on southwestern Indian tribes. II. Local genetic differentiation in the Papago. Am. J. Hum. Genetics 22:24-29. [ Links ]














