Introduction
Methicillin-resistantStaphylococcus aureus(MRSA) has been a common nosocomial pathogen and has become a major problem in hospitals worldwide. It produces a wide variety of virulence factors that contribute to its ability to colonize and cause a wide range of diseases such as folliculitis, conjunctivitis, cellulitis, deep abscesses, osteomyelitis, meningitis, sepsis, endocarditis and pneumonia.1,2,3Surveillance of MRSA infections has shown that there are two types of strains: healthcare-associated (HA-MRSA) or community-acquired (CA-MRSA) MRSA.4, 5The prevalence of MRSA in Mexico is high, with wide variations (24.1-80%).6A study carried in five hospitals of Monterrey, Nuevo León, Mexico, in 2013, detected 190 MRSA strains causing healthcare associated infection (HCAI).7Another study describes the first MRSA outbreak in a tertiary-care oncology hospital in Mexico City. The results showed that the source of this MRSA outbreak was a transferred female patient with a complicated bone-joint-prosthesis infection.8
MRSA strains are characterized by the presence of a mobile genetic element called staphylococcal chromosomal cassette (SCCmec); this element carries themecAgene that determines resistance to methicillin.9A small number of MRSA lineages have emerged from the transfer of SCCmecinto successful methicillin-susceptibleS. aureus(MSSA) clones. Using multilocus sequence typing (MLST), Enright and colleagues demonstrated that MRSA clones evolved from five different groups of related genotypes or clonal complexes, each arising from a distinct ancestral genotype.10The clonal framework of the MRSA population in different Mexican hospitals has been the subject of several studies. The ST5-MRSA-II-New York/Japan clone has been shown to be particularly successful in establishing itself in Mexico.7,8,11,12,13
The aim of this study was to identify international clones of MRSA circulating in theHospital Regional de Alta Especialidad de Veracruz, Mexico (HRV), and to track changes in their prevalence during the study period (September 2009 to September 2010).
Materials and methods
Hospital setting
The HRV in Veracruz, Mexico, is a 360-bed tertiary-care teaching hospital with 36 wards. The hospital serves the population of the Veracruz metropolitan area, which has 811 671 inhabitants. The hospital also serves the rest of the state of Veracruz (population 7 643 194) as well as three neighboring states: Oaxaca, Chiapas and Tabasco. During the study period, the hospital had 100 972 first inquiries, 19 768 emergency consultations, 85 066 specialty consultations and 15 414 hospitalizations.
Bacterial strains
A total of 107 MRSA isolates from individual patients were analyzed in the present study. The strains were collected between September 2009 and September 2010. Identification was done at Sensititre ARIS 2X automated system (Thermo Fisher Scientific Inc., Waltham, MA USA.). All MRSA isolated from invasive and non-invasive diseases during the study period were sent to the Department of Vaccine Evaluation for molecular typing.
Antimicrobial susceptibility tests
We used the Sensititre ARIS 2X automated system to perform antimicrobial susceptibility tests for amoxicillin, ampicillin, cefepime, cefotaxime, cefuroxime, cephalothin, chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, imipenem, meropenem, oxacillin, penicillin, rifampin, teicoplanin, tetracycline, trimethoprim-sulfamethoxazole and vancomycin, following the Clinical Laboratory Standards Institute guidelines.14
Molecular typing
The genomic DNA was prepared as described previously; after digestion with SmaI endonuclease, the DNA was separated using a CHEF-DR II system (Bio-Rad, Birmingham, UK).15The strains BK2464, HPV107, HDE288, and EMRSA-16, which represent the New York/Japan, Iberian, Pediatric and EMRSA-16-UK clones were included in each PFGE gel as controls. The control strains were kindly provided by Herminia de Lencastre from the Laboratory of Molecular Genetics of theInstituto de Tecnología Química e Biologica da Universidad Nova de Lisboa. The different clones were compared according to the criteria of Tenover.16Staphylococcal cassette chromosome mec element (SCCmec) typing and multilocus sequence typing (MLST) based on sequencing internal fragments of sevenS. aureushousekeeping genes used to define the allelic profile and a sequence type (ST), according to the MLST database (http://www.mlst.net) were performed as described previously.17,18
Computer fingerprint analysis
A computer analysis of the banding patterns obtained by PFGE was done using the Gel Compar II software, v. 6.6.11 (Applied Maths Inc.) after visual inspection. The reference strainS. aureusNCTC 8325 was included in each gel to normalize the PFGE profiles. The Dice coefficients were calculated to compute the similarity matrix and were then transformed into an agglomerative cluster by the unweighted pair group method with arithmetic average (UPGMA).
Results
MRSA isolates
Between September 2009 and September 2010, a total of 107 MRSA clinical isolates from theHospital Regional de Alta Especialidad de Veracruz, Mexico, were collected and analyzed. They were taken from 58 (54%) female patients and 49 (46%) male patients. The median age of the patients was 42 years (ranging from 1 to 80 years). The isolates were obtained from various body sites associated with S. aureus infections: blood (35; 33%), wound infections (33; 31%), bronchial aspirate (15; 14%), peritoneal fluid (7; 6%), cerebrospinal fluid (4; 4%), dialysis fluid (3; 3%) and other sources (10; 9%). The MRSA isolates were found in different wards: internal medicine (49; 46%), surgery (18; 17%), intensive care unit (19; 18%), traumatology (11; 10%), neurosurgery (4; 4%), emergency (2; 2%) and outpatient (4; 3%).
Antimicrobial susceptibility
All strains showed resistance (100%) to amoxicillin, ampicillin, cefepime, cefotaxime, cephalothin, clindamycin, erythromycin, imipenem, meropenem, cefuroxime, oxacillin and penicillin; some strains showed resistance to chloramphenicol (76%), gentamicin (16%), tetracycline (3%), trimethoprim/sulfamethoxazole (2%) and rifampicin (3%). No resistance was detected against vancomycin and teicoplanin.
Molecular typing
The PFGE pattern separated the MRSA strains into two clones: the New York/Japan clone (NY/J) and subtypes [NY/J28, NY/J35, NY/J31 and NY/J21] (97%) and the Iberian clone (IB), with three subtypes (IB1, IB2 and IB3 [3%]). The subtype NY28 was most abundant (75%). The results of the computer analysis of the banding patterns show a clear division between the two clone groups (NY/J and IB). The patterns of the NY/J clone and its subtypes had 79.7-88% similarity with the control strain BK2464 (representing the NY/J clone), while the profiles of the IB and subtypes showed 67.4-96.8% similarity with the control strain HPV107 (representing the IB clone) (figure 1). The NY/J clone was found in all wards of the hospital and was present throughout the study period. The IB clone was found only in the emergency (IB1) and intensive care units (IB2 and IB3) in December 2009, and April and May 2010, respectively. The SCCmec and MLST analysis showed the presence of two STs: ST247-I and ST5-II, related to the Iberian and New York/Japan clones, respectively.
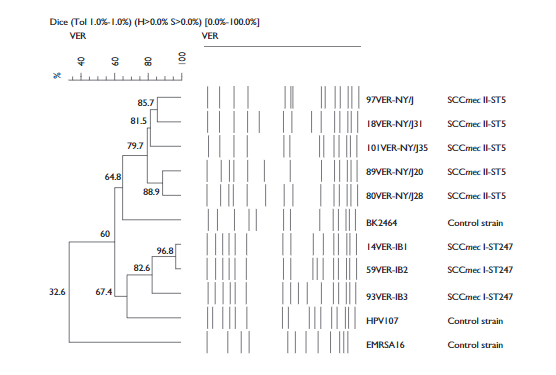
Figure 1 Dendrogram comparing MRSA clones (New York/Japan [NY/J] and Iberian [IB]) from the Hospital Regional de Alta Especialidad de Veracruz, Mexico, with different international MRSA clones (BK2464, HPV107 and EMRSA-16, represent the New York/Japan-USA, Iberian and EMRSA-16-UK clones respectively), as well as five subtypes of NY/J and three subtypes of IB. Patterns were clustered by the unweighted pair-group method using arithmetic averages, and the similarity coefficients were generated from a similarity matrix calculated with the Dice coefficient
Discussion
The wide availability of antibiotics and their indiscriminate use has exerted selective pressure on MRSA clones throughout the world.19,20The present study shows evidence of the presence of two pandemic clones of MRSA circulating in the HRV: the New York/Japan clone (ST5-SCCmec-II) and the Iberian clone (ST247-SCCmec-I). The New York/Japan clone, described by some studies as the most common clone in the United States of America (USA) and Japan,21has been described previously in other hospitals in Mexico.7,8,11,12,13However, this is the first report of this clone in the South of Mexico, which demonstrates its ability for geographical spreading, probably favored by its resistance and virulence factors. The importance of this finding lies in the fact that the first resistant strain of MRSA with vancomycin minimum inhibitory concentration (MIC) of 1024ug/ml belongs to the New York/Japan clone.22Furthermore, Khokhlova and colleagues reported the first case of fatal community-acquired necrotizing pneumonia caused by MRSA strains associated with the New York/Japan MRSA clone.23
The pandemic Iberian clone (ST247-I), which accounted for 3% of the isolates examined in the present study, has been reported as a common clone in Europe although there has been a decrease in the incidence of this clone in several European countries.10,24This is the first time that this clone has been detected in Mexico (three different subtypes; IB1 to IB3). Subtype IB1 was first isolated in the emergency ward from a patient with ear infection; the medical record showed that the infection had appeared after the patient returned from a travel to the USA. The MRSA strains of subtypes IB2 and IB3 were isolated later in the intensive care unit of the hospital from two more patients, showing that the Iberian clone had traveled from the external wards to the critical areas of the hospital. Previous studies have shown that the Iberian clone has spread widely in Europe and the USA, causing severe hospital outbreaks in these regions.25,26,27MRSA strains of the Iberian clone were the first to develop intermediate resistance to vancomycin (VISA) and reduced sensitivity to teicoplanin.28,29,30,31Glycopeptides are considered last-choice antibiotics against infections caused by MRSA and other Gram-positive microorganisms.
The antimicrobial resistance pattern to different antibiotics remained stable throughout the study period, similar to what has been observed in other studies.32,33All strains were resistant to 12 antibiotics and only some strains showed different resistance levels to five antibiotics, as shown in the results. The multi-resistance of the New York/Japan clone and the Iberian clone has been reported in other studies.34,35 However, our results showed that the Iberian clone strains (n=3) are susceptible to gentamicin and tetracycline, and resistant to rifampicin. The phenotype of the Iberian clone that showed resistance to rifampicin was found in clinical isolates of MRSA in 2004 in Spain, where the prevalence of this phenotype increased from 25% in 2004 to 45% in 2006. That study analyzed mutations of therpoBgene involved in the resistance to rifampicin; in this work an internal sequence of generpoB(432 bp) was amplified by PCR, this region includes the rifampicin resistance-determining; the PCR product was analyzed by DNA sequencing. The results showed mutational changes in 481His/Asn, 468Gln/Lys, 477Ala/Thr and 527Ile/Leu, the last three mutations associated with a high-level rifampicin resistance.36
Our study has several limitations. First, our collection is based on a small number of isolates collected in a short period of time. However, we do not believe there is a bias in the collection of isolates, because this was constant throughout the year of study. Second, prospective studies are needed to determine the incidence of infections due to MRSA in the HRV in order to implement measures aimed at preventing the dissemination of these clonal lineages. Despite this limitation, these data remain useful because they could be used primarily for monitoring trends of dissemination of these clones in Mexico, and could provide a basis for detection in other hospitals.
Several studies have shown that one or a few clones are predominant in each hospital, similar to what we found. In conclusion, this study establishes the presence of two very important clonal lineages of MRSA: New York/Japan and Iberian clone in hospital environment.











 nueva página del texto (beta)
nueva página del texto (beta)


