Introduction
Lung cancer is the most lethal malignant disease worldwide, it is expected that the number of cases of this disease will increase in the next few years, particularly in developing countries.1 Lung cancer has been classified in two major histopathological groups: small cell lung carcinomas (SCLC) and non-small cell lung carcinomas (NSCLC) that represents 80 to 85% of the total lung cancer cases. According to its histology, the NSCLC can be classified in: large cell carcinomas (LCC) 10%, squamous cell carcinomas (SCC) 40%, and adenocarcinomas (AD) 40%.2 NSCLC has also been grouped at the molecular level, according to a new genetic expression signature based on 42 differentially expressed genes (with a fold change >2.6) significant statistically (t test, P value <10−18, with FDR <10−16), highlighting in other propose genes: high molecular weight keratins (KRT), NKX2-1 (TITF1), TP63, and DSG3 (desmoglein 3), as a part of the new molecular classification for the human NSCLC-histological group.3 Nevertheless, in the pulmonary oncological field, clinical oncological prognoses and therapies are based on the relevant functional driver genes associated with the biology of lung cancer, particularly: EGFR-gene mutations, EML4-ALK-genetic fusions,4 and others oncogenic drivers as: BRAF, HER2, ROS1, RET, FGFR1, and PI3K, in human NSCLC.5
However, despite marginal advances in the diagnosis and oncological therapies, the clinical outcome and prognosis have room to improve significantly for patients. Therefore, the treatment specifically addressed to genetic-epigenetic targets for the applied lung translational oncological medicine is a priority for translational oncological medicine.
In this topic, high-performance epigenomic strategies have revealed a new picture of the complexity of the malignant cellular and molecular mechanisms of lung cancer, as well as their potential application as biomarkers, and NSCLC therapies.
Based on the definition of the National Cancer Institute (NCI, Dictionary of Cancer Terms), a biomarker is “a biomolecule identified in fluid-tissues as blood, and other body-fluids, as well as solid tissues, as a sign of a normal versus abnormal physiological condition and/or disease-processes”.6 The concept of Epigenetic-biomarkers has been incorporated in the personalized medicine as cancer-epigenetic drugs and/or epigenetic-target therapies. Therefore, tumor-biomarker functional and clinically may be used, as a determinant diagnostic and/or prognostic aggressive-malignancy guidance in cancer biology and clinical oncology research,7 where epigenetic biomarkers will be widely used in lung tumor biology, oncological prognosis, overall survival, early-late recurrence, and/or therapy response in human NSCLC.8,9,10
Epigenetics in NSCLC
Most cases of NSCLC exhibit genetic alterations that have been widely described; however, a small number of NSCLC cases have been identified without genetic mutations and/or chromosomal-genomic aberrations in the fundamental oncological driver genes. The study of the three levels of the human cancer epigenome (DNA methylation, histone code modifications and nucleosome positioning) has allowed us to propose new molecular pathways in the hereditary epigenetic marks that accompany transient-reversible and hereditary permanent transcriptional states under normal conditions, embryonic development, non-malignant diseases and scenarios of malignant diseases.11
The first lung cancer epigenetic studies carried out by Issa and colleagues and Baylin and Herman between 1996 and 1998, reported emerging epigenetic biomarkers (Estrogen Receptor Gene-Promoter Methylation) in both, environmental carcinogenic lung cancer mice models, and lung carcinomas from smokers and never-smokers patients.12 Some of which have been proposed as early epigenetic events (increased DNA methyl-transferase activity) associated with lung carcinogenesis promotion and primary lung tumor progression.13 Where DNA methylation aberrations on driver-Tumor Suppressor Genes (p16INK4a) have been proposed in NSCLC (lung precursor lesions hyperplasias, adenomas, AD and SCC) as early epigenetic biomarkers with potential using in lung cancer diagnosis.14,15In addition, two decades ago, several emerging epigenetic disorders have been described and classified at DNA methylation, covalent histone modifications, and nucleosome remodeling levels on several oncogenic driver-genes (p16, H-Cadherin, RARb, APC, MGMT, RASSF1A, CDKN2A, SHOX2, etc.), persistently associated to lung cancer diagnosis and prognosis.16
Moreover, in the last decade several oncological-research efforts have addressed large scale descriptive epigenetic studies, well-known as cancer epigenomics, into identify massive spreading altered epigenetic marks through -modulated-regulated- cancer epigenome, well known as functional epigenomics, into identify epigenetic-targets impacting on lung tumorigenesis, with its translational medicine implications.17 On that subject, some new proposed molecular mechanisms involved in lung cancer histopathological progression, prognosis and efficient oncological-therapeutic protocols, have been proposed in NSCLC patients, using others epigenetic-inhibitors as Histone Deacetylases-inhibitors including Vorinostat plus cisplatin and paclitaxel, consolidating an epigenetic therapeutic field in NSCLC.18
Epigenetics therapeutics in NSCLC
Based on the reversible nature of epigenetic modifications of methylated DNA and post-translational modifications of histones, some therapeutic strategies have been developed reversing malignancy in the cancer initiation, promotion and progression processes, consolidating the use of epigenetic biomarkers in the course and diagnosis of the malignant diseases.19,20,21
Three identifiable major molecular epigenetic patterns have been described at DNA epigenetic level: a) DNA global-hypomethylation, b) Region-specific DNA hypermethylation, and c) Genome-wide hypomethylation. These epigenetic aberration-modifications at epigenome-wide level, have led us to identify dysfunctional abilities on a) cellular-genetic expression patterns, b) physiological homeostasis of the cellular proliferation, and c) cellular-differentiation control. All of these cellular-disabilities, additionally respond to a higher functional complexity, affect chromatin remodeling complexes, nucleosome-assembling, histone code patterns, and epigenetic modulators epigenome-occupancy (figure 1).10 Some of these mechanisms have been proposed in lung-malignant diseases controlling transcription states, defining genetic expression levels and patterns, producing aberrations in the tumor-biomarker expression signatures through malignant tumorigenesis and histopathological progression compromising lung tissue at early and/or late NSCLC stages.16
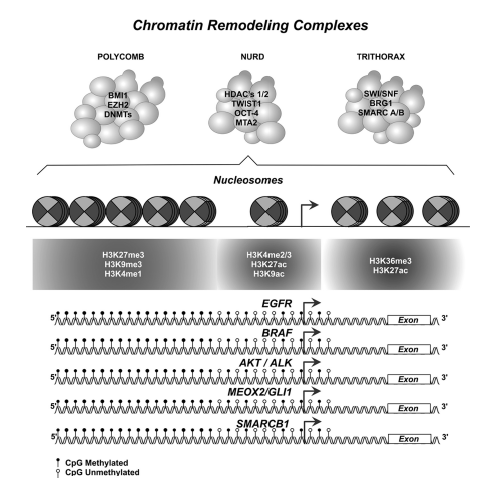
Figure 1 Epigenome depicture model: Chromatin remodeling complexes, nucleosome position, histone code patterns, and DNA methylation patterns in driver gene-promoter sequences.Probable epigenetic aberrations in modulated promoter-sequences of gene-drivers and/or epigenetic biomarkers functional-biological, and clinically involved in non-small cell lung carcinomas (NSCLC ), tumorigenesis, histopathological progression, prognosis and therapy response (additional information in table I and II).
Based on this background, targeted-epigenetic therapeutic protocols have been proposed as strategies that reactivate and/or regulate transcriptional activity, by inhibiting epigenetic enzimes,9 controlling oncogenes, tumor suppressor genes, and specific gene-signatures for each histopathological and molecular subtypes in NSCLC.3Thus, it is expected that in the near future there will be a better knowledge of the lung cancer epigenome (figure 1), and that these efforts will allow the development of therapies based on epigenetic drugs.17
Additional epigenetic-therapeutic strategies, have been based on the functional activity of the dynamic structure of chromatin, but centered on the enzymatic capacity of energy-dependent protein complexes leading to covalent post-translational modifications on the nucleosome, modulating several cellular processes, such as transcription, gene expression, as well as DNA recombination, replication and DNA damage repairing.22,23
In this regard, some investigations have been conducted, using azacitidine (30-40 mg/m2/d) plus Entinostat (7 mg on days 3 and 10, each 28-day cycle) in phase I-II clinical trials with NSCLC patients.24 On these studies, gene-promoter hypermethylation status has been proposed as a quantitative negative prognostic factor (P< 0.001), based on the quantifiable DNA-Methylation on driver gene-promoter sequences, as APC, RASSF1A, CDH13, and CDKN2A in stage I and III NSCLC patients.8
Additionally, in NSCLC, some mutations have been identified in genes codifying for the enzymatic subunits of the four classes of chromatin remodeling complexes (SWI/SNF, ISWI, CHD y INO80), such as ARID1A, BRG1, ARID2 and CHD7 (table I).25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48Other mechanisms of resistance identified include altered function of SMARCE1 subunits of the SWI/SNF1 group, which conduce to overexpression of EGFR and contributing in resistance to inhibitors of MET and ALK in NSCLC;49 the overexpression of BRG1 and BRM in the cisplatin toxicity,50 or doxorubicin chemo-resistance mechanisms associated with the expression of the subunits SMARCB1 and SMARCA4 (BRG1) in NSCLC cells.51 This findings suggest the importance of the three main epigenetic mechanisms, DNA methylation, histone code modifications and chromatin remodeling complexes in cancer progression (table I and II).52,53,54,55,56,57,58
Table I Alterations in chromatin remodeling complexes in NSCLC
| TRITHORAX Complex SWI/SNF Subunits | Gene involved and alteration | Study samples | Reference(s) | |||
| SMARCB1 | Mutated in lung AD patients, associated with wood smoke exposure: Mutation Frequency 73.7% (14/19). | Prospective cohort: patients diagnosed with lung AD with WSE from 2014-2017 at the Thoracic Oncology Clinic of the Instituto Nacional de Cancerología, México. n=19. | (25) | |||
| BRM (SMARCA2) | Decreased expression in NSCLC samples (6 of 60 samples, ~10%) and human NSCLC lines (6 of 20, ~30%). BRM acts as tumor suppressor protein in lung cancer. | NSCLC samples from lung tumor samples (Stage I-IIIA) banked at the Uni style="border-bottom:1pt solid black;"versity of North Caroline from 1997-1999, n=60. Human NSCLC cell lines derived from lung AD. n=20. | (26) | |||
| BRM (SMARCA2) | Altered cellular localization of BRM is a useful marker for NSCLC prognosis: Positive nuclear BRM localization is associated with a favorable prognosis in SCC and AD patients with 5 year-survival (53.5%) Membranous BRM localization is associated with a poorer prognosis in AD patients with 5 year-survival (16.7%). | Tissue microarray composed of 300 NSCLC cases selected from the Armed Forces Institute of Pathology Archive, Washington, DC. n=150 AD. n=150 SCC. | (27) | |||
| BRG1 (SMARCA4) | Concomitant loss of BRG1/BRM expression is a poor prognostic indicator in NSCLC patients. Nuclear Co-expression of BRG1/BRM correlates with a better prognosis in NSCLC. | Prospective cohort: median 36-month follow-up period. Tumor samples banked at the University of North Carolina. The specimens were derived from patients with stage I–IIIA, NSCLC. n= 6 patients with BRG1/BRM-negative tumors. n= 54 patients with BRG1-positive tumors with stage I, II, and III. NSCLC cases selected from the Armed Forces Institute of Pathology Archive, Washington, DC. n=15 of 28 M-BRM positive NSCLC. | (26, 27) | |||
| BRG1 (SMARCA4) | SMARCA4 gene promoter hypermethylation does not occur in primary lung tumors or cancer cell lines. Somatic point mutations of the SMARCA4 gene are present in a small subset of lung tumors and lung cancer cell lines. | NSCLC patients at The Johns Hopkins University Scholl of Medicine. n= 20 lung primary tumors with LOH on Chr:19p. n=52 lung primary tumors. Human lung cancer cell lines from American Type Culture Collection (ATCC, Rockville, MD) n=10. | (28, 31) | |||
| BRG1 (SMARCA4) | Decreased expression in lung cancer cell lines and lung primary tumors. Genetic expression restoration of BRG1 in H1299 lung cancer cell line, identified BRG1 target genes (CYP34A and ZNF185), using cDNA microarray analysis. | Tumor tissues from 27 lung cancer patients, provided by the CNIO Tumor Bank Network, (Madrid, Spain), by collaboration with the Hospital Universitario 12 de Octubre. n=13 SCC. n=17 AD. H1299 lung cancer cells from ATCC and cDNA microarray (CNIO OncoChip). | (32) | |||
| BRG1 (SMARCA4) | BRG1 loss altered cellular morphology and increased tumorigenic potential in NSCLC. Inactivation of BRG1 in NSCLC were associated with variations in chromatin structure, including differences in nucleosome positioning and occupancy surrounding transcriptional start sites of key cancer-associated genes. | Human NSCLC and NSCLC cell lines from the ATCC. Agilent micrroarray analysis. | (33) | |||
| BRG1 (SMARCA4) | Reduced expression of SMARCA4 was associated with poor prognostic and overall survival. Lower expressed SMARCA4/BRG1 associated with increased benefit from cisplatin-based chemotherapy in resectable NSCLCs. SMARCA4-decificient lung AD shows a morphological diversity and genotypic spectrum, with absence of expression of TTF1 in the presence of expression of HepPar-1, and absence of EGFR-driver mutations. These features should be recognized as a distinct new subtype of NSCLC. | Patient cohorts: The Director´s Challenge Study, University of Michigan and clinical outcomes in NSCLC from multiple institutions, Memorial Sloan-Kettering Cancer Center, the H. Lee Moffitt Cancer Center and Research Institute, the Dana-Ferber Cancer Institute, and the National Center Institute of Canada Clinical Trials Group. n=440. | (34) | |||
| NSCLC cases diagnosed at the Institute of Pathology, University Hospital of Erlangen, Germany. n=20. | (35) | |||||
| BAZ1B (WSTF) | WSTF is an oncoprotein in lung cancer, its overexpression promotes proliferation, colony formation, migration and invasion of lung cancer cell lines A549 and H1299. WSTF overexpression also promotes tumor growth of lung cancer cells in mouse xenograft models. | Human lung cancer cell lines: A549, and H1299 from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. Male athymic nude mice with four weeks old. | (36) | |||
| NuRD Complex CHD Subunits | Gene involved and alteration | Study samples | Reference(s) | |||
| RBBP7 (RbAp46) | RbAp46 is a prognostic biomarker in NSCLC and involved in lung cancer cellular migration. | Cancer cell lines stablished from a 64- year-old man with a poorly differentiated lung AD (CL1-0 and CL1-5). Department of Internal Medicine National Taiwan University Hospital, Taipei, Taiwan, Republic of China. Paired surgically resected lung carcinomas, clinical staging I-IV from patients of Chang Gung Memorial Hospital: n=106 AD. n= 48 SCC. | (37) | |||
| MTA2 | MTA2 are distributed in both nuclei and cytoplasm in NSCLC cells. Nuclear MTA2 distribution was detected in 66.4% (n=148) of NSCLC cases, and was correlated with advanced TNM stage, tumor size, and lymph node metastasis. Cytoplasmic MTA2 status was not associated by age, gender, tumor stage, histology, grade, and lymph node metastasis. Nuclear MTA2 expression is correlated with poor overall survival. | Patient cohort’s NSCLC tissue cases obtained between 2000 and 2005 from Hunan Providence People´s Hospital, China. n=223. NSCLC cell lines n=2. | (38) | |||
| HDAC1 | HDAC1 expression is associated with the histopathological progression and prognosis of lung cancer. HDAC1 is highly expressed in NSCLC cell lines, and HDAC1 knockdown inhibits cellular invasion, inducing apoptosis in NSCLC cells. | Meta-analysis study. Original articles in Embase, Web of Science, PubMed. December 2016 | (39) | |||
| Human NSCLC cell lines A549, H1299 and LK2 from ATCC (Manassas, VA, USA). | (40) | |||||
| HDAC2 | Overexpression of HDAC2 in lung cancer tissues. HDAC2 has oncogenic properties in human lung cancer cell lines. | Human NSCLC cell lines A549, NCI-H358, and NCI-H460 from ATCC. | (41, 42) | |||
| CHD4 (Mi-2β) | Young never-smoker patients with lung AD harbored germline mutations in CHD4 (rs74790047, p.D140E). The CHD4 susceptibility loci has been identified to be associated with lung cancer risk. | Cohort never-smoker Chinese patients diagnosed with lung AD, at 45 years or younger. From West China Hospital From 2011 to 2016. n=36. | (43) | |||
| POLYCOMB Complex Polycomb Repressive Complex (PRC) | Gene involved and alteration | Study samples | Reference(s) | |||
| PRC2/ Subunit Enhancer Zeste Homolog 2 (EZH2) | Deletion and inactivating mutations of EZH2 gene is present in 14% (33/230) of human ADC samples. EZH2 gain/amplification is prevalent in 42% (97/230) of ADCs. | Human lung AD samples from Cancer Genome Atlas Research (TCGA). n= 230. | (44) | |||
| PRC2/ Subunit Enhancer Zeste Homolog 2 (EZH2) | EZH2 is highly expressed in SCCs and NSCLC brain metastases. EZH2 expression correlated with tumor progression and prognosis of NSCLC | Cohort from patients who underwent surgical resection between 1999 and 2006 at The University of Texas MD Anderson Cancer Center (Houston, Texas). n= 221 SCC, n= 320 AD n= 36 NSCLC (9 SCCs and 27 AD) with brain metastasis. | (45) | |||
| PRC2/ Subunit Enhancer Zeste Homolog 2 (EZH2) | EZH2 promotes lung cancer progression through transcriptional repression of the metastasis suppressor gene TIMP-3 in NSCLC. | Cohort from patients with primary NSCLC diagnosed at the Department of Thoracic Surgery of Nanjing Chest Hospital between 2005 and 2008. n= 60. | (46) | |||
| PRC1/ Subunit BMI-1 | BMI1 is overexpressed in stage III and IV tumors. BMI overexpression has been associated with progression of NSCLCs. | Human tumor samples NSCLC from 1st Department of Surgery, Medical Faculty, Palacky University, Olomouc, Czech Republic, between 1996 and 2001. n= 179: n= 104 (stage IIIa), n=20 (stage IIIb) and n=21 (stage IV). | (47) | |||
| PRC1/ Subunit BMI-1 | BMI1 is overexpressed in lung AD samples, and correlated with clinical features of lung cancer, including metastasis rates. Knockdown of BMI1 reduced cellular migration and invasion/metastasis of A549 and SPCA1 lung AD cell lines, further upregulated metastasis gene (PTEN) and downregulated pAKT and VEGF expression in lung AD cells. | Lung Cancer Samples of Chinese Patients diagnosed in 2009 and 2010 at the Dalian fifth Hospital, Dalian, China. n= 38: 18 men and 20 women, ranging 41-79 years old. Lung cancer cell lines: A549 and SPC-A1 | (48) | |||
SWI/SNF: Switch/Sucrose Non Fermentable; SMARCB1: SWI/SNF Related, Matrix Associated Actin Dependent Regulator Of Chromatin, Subfamily B, Member 1; SMARCA2: SWI/SNF Related, Matrix Associated Actin Dependent Regulator Of Chromatin, Subfamily A, Member 2; SMARCA4: SWI/SNF Related, Matrix Associated Actin Dependent Regulator Of Chromatin, Subfamily A, Member 4; BZ1B (WSTF): Bromodomain Adjacent To Zinc Finger Domain 1B; RBBP7: RB Binding Protein 7, Chromatin Remodeling Factor; MTA2: Expression of metastasis-associated protein 2; HDAC1: Histone deacetylase 1; HDAC2: Histone deacetylase 2; CHD: Chromodomain Helicase DNA Binding Protein; EZH2: Enhancer Zeste Homolog 2; PRC1 and PRC2: Polycomb Repressive Complex 1 and 2; BMI1: BMI1 Proto-Oncogene, Polycomb Ring Finger; AD: lung adenocarcinoma; WSE: Wood smoke exposure; NSCLC: Non-Small Cell Lung Cancer; SCC: Squamous Cell Carcinoma; M-BRM: membranous BRM staining; ATCC: American Type Culture Collection; CNIO: Spanish National Cancer Centre.
Table II Histone profile aberrations in NSCLC
| Histones | Gene involved and alteration | Study samples | Reference(s) | |||
| H3K4me3/H3K27AC | Sonic Hedgehog GLI-1 Overexpression, Chemoresistance and Poor Overall Survival. | A total of 123 tumor samples from 2 cohorts of NSCLC, (33) INER and (90) INCAN. NSCLC cell lines A427, A549, NH2347, HCC827 and H1975. | (52) | |||
| H3K4me3/H3K9me3 | Epigenetic (Histones) reprogramming at GLI-1 gene promoter sequences, by cancer drugs (cisplatinum) exposure. | NSCLC cell lines A427, A549, NH2347, HCC827 and H1975 | (53) | |||
| H3K4me3 | miR-375 Overexpression. | A549 lung adenocarcinoma cell line without NE differentiation and a typical SCLC cell line, ACC-LC-172. | (54) | |||
| Activated by ASH1 and Inhibits YAP1 in a Lineage Dependent Manner in Lung Cancer. | ||||||
| H3K4me3 | NFE2L3, Overexpressed. ETV4, Overexpressed. PRTG, Overexpressed. TMEM86A, Overexpressed. | 26 lung adenocarcinoma cell lines | (55) | |||
| Epigenetic Biomarkers of Lung Adenocarcinoma through Multi-Omics Data. | ||||||
| H3K27me3/H3K27AC | TWIST1/MEOX2, Overexpression. | 55 lung tumors (LT), 15 adjacent non-involved lung cancer matched tissues (LNAT) and 20 lung precursor lesions (LP) from Fresh Frozen (FF) and Formalin-Fixed and Paraffin Embedded (FFPE). | (56) | |||
| MEOX2 and TWIST1 Genes Are Associated with H3K27me3 Levels, Chemoresistance and Poor Prognosis in Lung Cancer. | ||||||
| H3K9me3 | Large Intergenic Non-Coding RNA (LincRNA) H19, Downregulation. | A549 Lung adenocarcinoma cell line | (57) | |||
| Mineral dust-induced genes (mdig), induce significant reduction of the H3K9me3 on H19 gene promoter. | ||||||
| Increased mdig and H19 correlate with aberrant heterochromatin and poorer survival in lung cancer patients. | ||||||
| H3K9me3 | Jumonji C (JmjC) Domain Proteins Family. Jmjd2C (GASC1, gene), Overexpressed. | 13 Lung AD, and 23 SCC. | (58) | |||
| Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. | ||||||
H3K9me3: tri-methyl lysine 9 of histone H3; H3K27AC: acethyl lysine 27 of histone H3; H3K4me3: tri-methyl lysine 4 of histone H3; H3K27me3: tri-methyl lysine 9 of histone H3; GLI1: Glioma-Associated Oncogene Homolog 1; ASH1: Like Histone Lysine Methyltransferase; YAP1: Yes Associated Protein 1; NFE2L3: Nuclear Factor, Erythroid 2 Like 3; ETV4: Ets Variant Gene 4; E1A: Enhancer-Binding Protein E1AF; PRTG: Immunoglobulin Superfamily, DCC Subclass, Member 5; TMEM86A: Transmembrane Protein 86A; MEOX2: Mesenchyme Homeobox 2; Twist1: Twist Family BHLH Transcription Factor 1; GASC1: Lysine Demethylase 4C.
Along with, overexpression of BRG1 and BRM in the cisplatin toxicity,50 or doxorubicin chemo resistance mechanisms associated with the expression of the subunits SMARCB1 and SMARCA4 (BRG1) in front to NSCLC cells.51 All of that, suggesting the importance of the three main epigenetic mechanisms DNA methylation, histone code modifications and chromatin remodeling complexes in cancer progression and potential new oncological therapeutic strategies in NSCLC, all of which have been shown in table I and II.
DNA gene promoters methylation in NSCLC
Based all above-mentioned, in NSCLC cells as well NSCLC solid tumors, several epigenetic spreadable alterations-aberrations have consistently been described, highlighting genome-wide DNA hypomethylation, and gene-promoter specific sequences DNA-hypermethylation.59,60
Alterations in DNA-sequences methylation located at 5’ position in the cytosine base in a CpG dinucleotide context, develop a pathophysiological condition in cancer cells known as hypermethylated “CpG islands”, in almost all NSCLC cases, these CpG islands are located mainly in tumor suppressor genes (Eg., O6-methylguanine-DNA-mehtyltransferase MGMT) codifying for a DNA-repair protein, which has a pivotal role for DNA-repairing, and cellular cycle control mechanisms.61 Thus, DNA hypermethylation at specific gene promoter sequences represents an important mechanism for loss of genetic function for several NSCLC histopathological types. In accord to two-hit model from Knudson, the phenotypic consequence of loss of the tumor-suppressor gene function, is not seen unless both alleles of the tumor suppressor genes are inactivated. In NSCLC there have been described gene mutations for one genetic allele, meanwhile, an additional allele is epigenetic silenced by hypermethylation. As functionally occurs for O6-MGMT protein removing carcinogen-induced O6-methylguanine adducts, resulting in G-A transition mutations in several driver-genes, such as p53 (P53) and K-RAS, promoting malignant abilities.61 In advanced NSCLC epigenetic inactivation events by DNA methylation precede to RAS genetic alterations, as well as in lung pre-malignant lesions, with a significant impaired overall survival in NSCLC patients.62
In addition, tumor suppressor genes epigenetically disrupted are often found in genome-wide regions, near of chromosomal deletions; whose genetic deletions cause loss of heterozygosity (LOH). On that epigenetic inactivation of important genes as, RASSF1A,63 and hypermethylated in cancer 1 (HIC1) gene (encoding a Zinc Finger Transcription Factor, at Chr:17p13.3) are usually inactivated by hypermethylation mechanisms in several lung malignant diseases.64 Nevertheless, recently it was proposed that higher-frequency LOH regions not caused by genetic-mutation mechanism, is a principal candidate tumor-suppressor gene disability mechanism, where no-additional mutated genes have nearly been found.30 However, 5-methylcytosine as per se known mutagenic nature, has been involved promoting tumorigenicity (G-T transversion mutations), caused by spontaneous hydrolytic deamination induced by tobacco carcinogens (benzo(a)pyrene diol epoxide), associated with tobacco consumption background in NSCLC patients.65
At least 50% of all inactivating point-mutations at coding region of the human TP53 tumor-suppressor gene, occur at methylated cytosines where methylated CpG dinucleotides increase UV-induced mutations, due that the methyl-group shifts the UV absorption spectrum for cytosine-base by the light-spectrum in sunlight exposure, affecting chemical and epigenetically gene-drivers in smokers with lung cancer.61,65
While, at epigenome-wide level have been identified several epigenetic aberrations in genome-loci, and/or focal DNA gene-promoters sequences as potential epigenetic biomarkers, highlighting homeobox-related (HOX) locus, known as HOX-clusters A, B, C and D encoding genes located at Chr:2, Chr:7, Chr:12 and Chr:17, epigenetically regulated by DNA methylation in both NSCLC cells and NSCLC tumors derived from stage I patients, highlighting promoter-sequences hypermethylation on HOXA7 and HOXA9 genes.59 As well as, DNA hypermethylation in additional HOX encoding-gene promoters as: OTX1, OSR1, IRX2 and NR2E1 for stage I-III NSCLC patients,29 reinforcing that DNA-methylation may be used as persistent epigenetic biomarkers for basic research and clinical-stage progression of NSCLC patients.60,66Finally, resulting in a) the activation of genetic repetitive elements, b) genomic DNA-instability and c) constitutive genetic expression of specific oncogenes patterns, representing pivotal key elements involved in carcinogenic and/or tumorigenesis processes, as part of the emerging hallmarks of cancer, as genome-instability phenomena, and probably epigenome instability-variability as emerging hallmarks of NSCLC.67
Histone code modifications in NSCLC
Epigenetic (post-translational) modifications of the histone code conforming to nucleosomes are continually contributing with reversible stability into the biological processes of transcription, nuclear architecture, and genomic stability, which occur through the N-terminal domain modifications by acetylation, methylation, phosphorylation, ubiquitination, sumoylation, ADP-ribosylation and deamination.68
On that epigenetic marks, by acetylation in lysine residues (Eg., H4K16ac) and trimethylation of the lysine 20 residue (H4K20me3), have importantly been identified as early lung epigenetic biomarkers. Importantly, loss of the differentiation-specific histone marker H3K9me3, continuously occurs in lower density CpGs regions in several types of malignant diseases, next to the gene promoter sequences known as “CpG island-shores”. Moreover, it has well been described that variation in the DNA methylation rates outside of the CpG islands, contributes directly to the NSCLC heterogenicity. Furthermore, it has been well characterized loss of the active histone marks H3K9ac and H3K4me3, with an increased enrichment of the repressive histone marks H3K27me3 and H3K9me2/me3 (figure 1), of which biological and clinically correlate with lung malignant progression, and poor therapy responses in NSCLC.16,69,70
Reports have described that low levels of H3K9me2 are associated with poor prognosis in lung cancer, and other epithelial types of cancer. While low levels of H3K18ac and H3K4me2 predict poor prognosis in NSCLC patients.21 In particular, Van Den Broeck and colleagues in 2008, have identified that trimethylation state in lysine 20 of histone H4 (H4K20me3) allows the prognosis stratification of AD-NSCLC patients with higher risk of death in clinical stage I. H4K20me3 has additionally been proposed as biomarker in subgroups of AD patients with poor differential clinical prognosis, and potentially determinant for the use of adjuvant chemotherapy.71
Additionally, while low levels of the histone mark H3K27me3 have been associated with chemo resistance and poor prognosis in NSCLC patients,56 additional reports have confirmed that higher expression of H3K27me3 correlates with better overall survival (OS) and better prognosis, redefining subgroups of NSCLC patients with an epigenetic phenotype and different clinical outcome.72
Additionally, despite histone marks H3K4me2, H2AK5ac and H3K9ac are related to global and disease-free survival in early clinical stages of NSCLC patients, undergoing curative surgical resection.73 However, due that histone modifications are associated with tumor suppressor gene repression mechanisms or oncogene activation, the behavior of such reversible histone code modifications must be studied in a depth manner, which will be necessary for a deep understanding to define their predictive role and/or functional in the experimental or clinical management in the NSCLC progression, and new pharmaco-epigenetics therapy field on NSCLC.
Conclusions
Future directions: Integrative studies by chromatin remodeling complexes in NSCLC
Over-represented emerging epigenetic biomarkers as biomolecular signals of the malignant cellular transformation processes represent powerful molecular tools in the transitory of molecular and histological steps, in accord with early diagnosis and/or late predictive tumor aggressiveness to predict therapy responses. To date remaining functionally unknown several epigenetic complex mechanisms to support novel applied epigenetic-drugs (pharmaco-epigenetics therapy) knowledge in NSCLC.
Spite that CpGs islands, represent approximately 1% of the total of human genome, located near of, or in gene-promoter sequences, global spread epigenetic marks in NSCLC epigenome has not just commonly been associated with the genome instability, and/or aberrant genetic expression patterns in early and late events in human NSCLC tumorigenesis. Instead or in addition chromatin remodeling complexes (Trithorax, NuRD, and Polycomb complexes) are epigenetically controlling genetic sequences in an active-dependent, but in an independent well-known DNA methylation status at several driver-gene promoter sequences, in others p16, H-cadherin, RASSF1A, APC, DAPK1, EGFR, BRAF, AKT/ALK, MEOX2/GLI1, SMARCB1, etc. (figure 1). Due that aberrant DNA methylation, histone code modifications and chromatin remodeling complexes gene expression and function rates, must be, begun as useful and standard tools in NSCLC diagnosis, lung malignant histological-phenotype subtype, clinical stage, malignant aggressiveness and therapy prognosis response, as well as, probably epigenetic-biomarkers associated with the risk for lung cancer development.16
However, to date relevant knowledge about higher molecular complexity of the epigenetic mechanisms, must be considered, but based on the several new molecular alterations of the well-assembled mechanisms of the Chromatin Remodeling Complexes, such as: Trithorax (SMARCB1, SMARCA2, SMARCA4, etc.), NuRD (RBBP7, MTA2, HDAC1, etc.), and Polycomb (EZH2, BMI-1, etc.) complexes (figure 1). Some of which have been clinically and oncologically validated on in vitro experimental NSCLC cellular and in vivo models, and implicated in NSCLC patients, described and summarized in table I. Which, in the advanced therapeutically strategies must be functional analyzed in synchronicity with the nucleosome structures and histone-code patterns on the functional epigenome-occupancy, whose biological and clinical implications in human NSCLC, have previously been reported, and summarized in table II. All of that, must be deeply studied, for a better and integral epigenetic-knowledge (DNA methylation, histone modifications and remodeling complexes patterns) into the control mechanisms of transcriptional regulations status, helpful to explain genetic-epigenetic cancer biomarkers expression (figure 1), all necessary for future concerns in lung cancer biology, personalized medicine, and emerging hallmarks of cancer applied into the translational medicine, accompanied by imaging technologies for diagnosis, and prognosis in NSCLC oncological therapies.











 text new page (beta)
text new page (beta)


