Introduction
A recent taxonomic description of the genus Klebsiella spp. includes three taxonomic subspecies; K. pneumoniae subsp. pneumoniae (KpI), K. pneumoniae subsp. rhinoscleromatis and K. pneumoniae subsp. ozaenae and our novel species; Klebsiella variicola (KpIII), K. singaporensis, K. michiganensis and K. quasipneumoniae (KpII), which has two subspecies: Klebsiella quasipneumoniae subsp. quasipneumoniae (KpII-A) and K. quasipneumoniae subsp. similipneumoniae (KpII-B).1 For these bacterial species novel genome sequences have been described from the strains 07A044, 18A0691 and FIHV2014.2 The misclassification of K. variicola and Klebsiella spp. genomes was first identified by whole-genome and rpoB phylogenies.3,4 Subsequently by K. quasipneumoniae and other K. variicola genomes the misclassification was evidenced by Average Nucleotide Identity (ANI).5 The aim of this work was detect misclassified K. variicola and K. quasipneumoniae genomes. We used an rpoB gene phylogenetic analysis and the results were validated by ANI. We identified the isolates misclassified previously described5 and identified new isolates that corresponded to K. variicola and K. quasipneumoniae that were deposited as K. pneumoniae. Likewise, we identified one K. variicola genome misclassified that corresponds to K. quasipneumoniae subsp. similipneumoniae.
Materials and methods
Identification of the K. variicola and K. quasipneumoniae genomes
The complete nucleotide sequence of rpoB (KVR801 v1_470024) gene from K. variicola 801 (CDMV00000000) was downloaded from the European Nucleotide Archive database.6 The rpoB sequence was analyzed by the BLASTn option (with default values) against 419 complete and draft Klebsiella spp. (tax id: 570) genomes deposited in GenBank database (last analysis 13/07/2015).7 The rpoB genes from Escherichia coli K-12-MG1655, Salmonella enterica Ty21a, Klebsiella oxytoca KCTC1686, K. quasipneumoniae subsp. quasipneumoniae 18A069 (KpII-A), K. quasipneumoniae subsp. quasipneumoniae 01A030 (KpII-A) and K. quasipneumoniae subsp. similipneumoniae 07A044 (KpII-B) genomes1 were included in the phylogenetic analysis. Subsequently, the rpoB nucleotide sequences of K. pneumoniae genomes with 100% nucleotide identity were filtered out to exclude a possible bias. All strain genomes identified as K. variicola and K. quasipneumoniae were included. Finally, a phylogenetic reconstruction was performed using the maximum-likelihood method with a Tamura-Nei-parameter model and 1000 bootstrap replications (Mega v6.06).8
The ANI was determined using the Average Nucleotide Identity Calculator9 the default parameters. In total, thirty-one K. variicola, nine K. quasipneumoniae and two K. pneumonia genomes were downloaded from GenBank and analyzed (table I). K. variicola At-22 (CP001891), K. quasipneumoniae subsp. quasipneumoniae 18A069 (CBZM000000000), K. quasipneumoniae subsp. similipneumoniae 07A044 (CBZR000000000) and K. pneumoniae MGH78578 (CP000647) were used as reference genomes of the respective bacterial species.
Table I Average nucleotide identity (ANI) between bacterial genomes from all isolates as queries, and K. variicola, K. pneumoniae and K. quasipneumoniae reference strain genomes
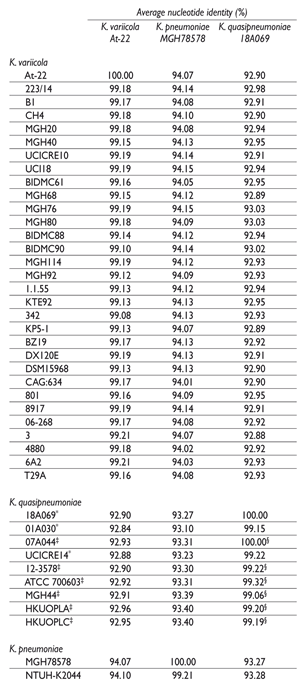
* Klebsiella quasipneumoniae subsp. quasipneumoniae
‡ Klebsiella quasipneumoniae subsp. similipneumoniae
§ K. quasipneumoniae subsp. similipneumoniae 07A044 used as reference genome in ANI calculation
Results and discussion
Klebsiella spp. genomes that corresponded to K. variicola and K. quasipneumoniae
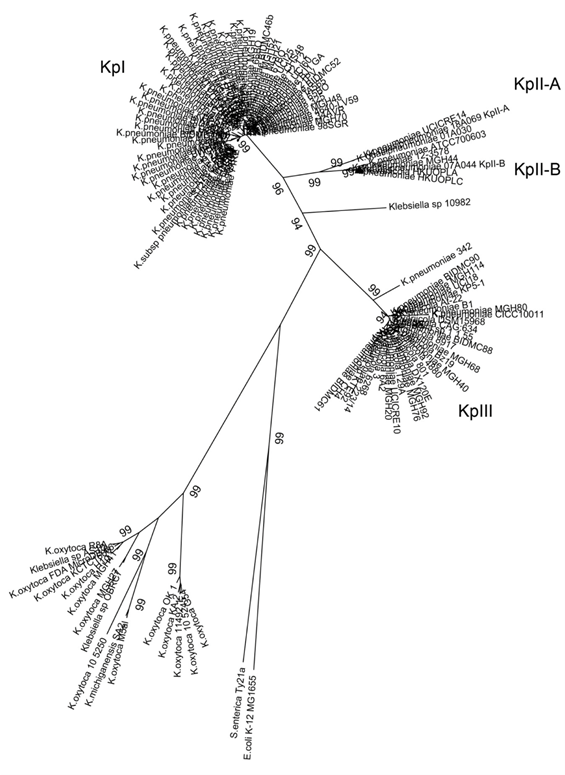
Both K. variicola as K. pneumoniae and K. quasipneumoniae isolates exhibit very similar biochemical features,1 and to date there are no reported biochemical tests for their appropriate differentiation.3,4,10,11 The phylogenetic analysis of the rpoB gene is more adequate than the 16S rRNA for Klebsiella spp. differentiation.12 Likewise, K. variicola DX120E was identified both by 16S rRNA as rpoB genes, however, a better resolution was obtained with rpoB gene analysis.13 In addition, K. variicola described in animals and plants was also included in the rpoB phylogenetic analysis.3 Previously concatenated housekeeping genes were proposed to differentiate of Klebsiella species.1,3 With this criterium K. pneumoniae 342 from maize,14 and K. pneumoniae KP5-1 from insect15 were found to be misclassified as K. pneumoniae.3 Actually the new K. variicola and K. pneumoniae isolates may now be properly identified by PCR. The first multiplex PCR, M-PCR-1 based on unique genes from each species would allow the differentiation in clinical laboratories of K. variicola and K. pneumoniae bacterial species.3 In the present study, the nucleotide BLAST analyses of rpoB gene sequence allowed the analysis of 419 Klebsiella spp. Genomes. All those strains with rpoB genes with 100% nucleotide identity to rpoB of K. pneumoniae MGH78578 were not further considered, as they had a correct taxonomic affiliation. 144 non-redundant DNA sequences were used for a phylogenetic analysis, showing seventeen putative K. pneumoniae strains corresponding to K. variicola bacterial species (figure 1). Their reported identification was K. pneumoniae MGH20, MGH40, MGH68, MGH76, MGH80, MGH92, MGH114, UCI18, BIDMC61, BIDMC88, BIDMC90, 223/14, KTE92, UCICRE10, B1, and CH4 and Klebsiella spp. 1.1.55. The Klebsiella spp. 1.1.55 clinical isolate was previously identified as a possible K. variicola.4 In addition, K. pneumoniae UCICRE14 corresponded to K. quasipneumoniae subsp. quasipneumoniae (KpII-A), whereas the K. pneumoniae 12-3578, ATCC 700603, MGH44 and HKUOPLC and K. variicola HKUOPLA corresponded to K. quasipneumoniae subsp. similipneumoniae (KpII-B) (figure 1).
Validation by average nucleotide identity analysis
The results obtained by phylogenetic analysis of rpoB gene were confirmed by determining the ANI value (table I). K. variicola At-22, K. quasipneumoniae 18A069, K. quasipneumoniae subsp. similipneumoniae 07A044 and K. pneumoniae MGH78578 were used as reference genomes in the ANI platform. This platform may use draft or complete genomes.7 When strains from the same subspecies were compared, ANI was >99%. Among the K. variicola genomes, an ANI value of >99.08 was obtained; similarly, an ANI value of >99.15% for the K. quasipneumoniae genomes was obtained. For the K. pneumoniae and Klebsiella spp. genomes that were identified as K. variicola in the present study, ANI values were <94.01% compared with those of K. pneumoniae MGH-78578. In contrast, an ANI value of >98.89% was obtained when K. variicola isolates were compared to K. variicola At-22 (table I). Previously, Chen and colleagues 5 detected nine misclassified K. variicola and four K. quasipneumoniae genomes. This update of Klebsiella genomes highlight that not only K. pneumoniae are misclassified, but also, K. variicola.
Not only K. pneumoniae and K. variicola but also K. quasipneumoniae isolates are not differentiated by biochemical methods. To address this problem, Brisse and colleagues1 proposed a phylogenetic analysis based on five protein-coding genes (rpoB, fusA, gapA, gyrA, and leuS) that clearly differentiates K. variicola and K. quasipneumoniae from K. pneumoniae bacterial species. Nevertheless, we previously showed that the rpoB nucleotide partial gene sequence (501 bases) also supports the differentiation of K. variicola and K. pneumoniae.3 This work confirms the correct taxonomic affiliation using the complete rpoB gene sequence.
General characteristics of K. variicola and K. quasipneumoniae strains and genomes
Misclassified K. pneumoniae and K. variicola genomes, now know to corresponded to K. variicola or K. quasipneumoniae, were submitted to GenBank in 2009. Other misclassified strains, such as Klebsiella spp. 1.1.55, and K. pneumoniae 342 genomes were deposited in 2008 (table II). The latter was reported to correspond to K. variicola in subsequent studies.1,3,16 Genome submissions were mainly to USA, Mexico and Malaysia, with the USA as the major contributor (table II). These isolates were obtained from many different sources, such as humans (from urine, pus bile and other organic tisues), giant pandas (feces), different sites of plants, and from diverse insect species (table II). The majority of bacterial genomes were submitted as drafts with 12 to 400 contigs, using mostly Illumina technology. Five K. variicola genomes were completely sequenced using the Sanger shotgun, PacBio and 454 technologies (completed by Sanger sequencing of gaps), and the draft genomes contained from 24 to 464 contigs.
Table II Characteristics of K. variicola, K. quasipneumoniae and K. pneumoniae genomes considered in the study
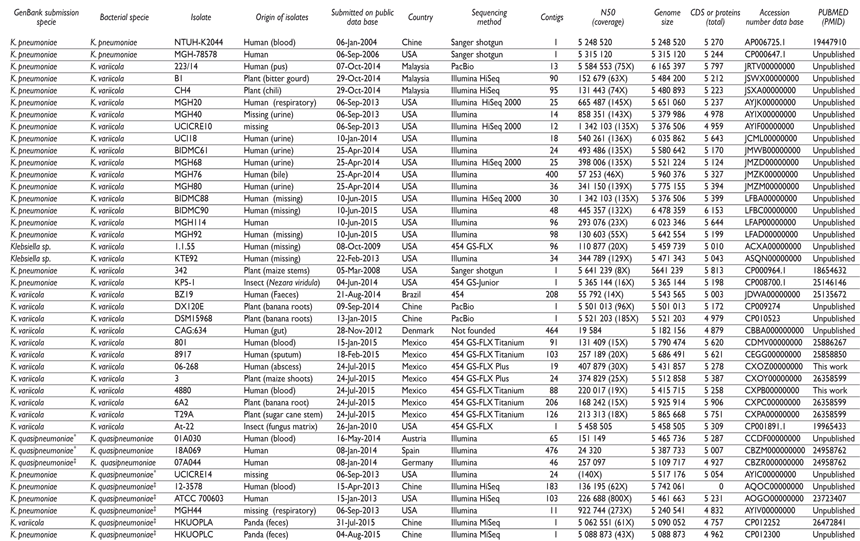
* Klebsiella quasipneumoniae subsp. quasipneumoniae
‡ Klebsiella quasipneumoniae subsp. similipneumoniae
The K. quasipneumoniae genomes were submitted in 2013 and 2015 from different countries, including Austria, China, Germany, Spain and USA. All of these genomes were isolated from humans and were sequenced using the Illumina platform, with contigs from 1 to 476 and genome sizes >5 Mb and a content of CDS or proteins of 4 757 to 5 287 (table II).
Final considerations
It is a fact that Klebsiella spp. isolates have been misclassified. Several isolates of both K. variicola and K. quasipneumoniae from various sources were identified as K. pneumoniae. We report here, on a genomic basis, on misclassified K. pneumoniae, K. quasipneumoniae and K. variicola isolates from humans, animals, plants and insects. Nevertheless, the recent use of different methods such as PCR, phylogenetic analysis of rpoB gene and ANI for the proper differentiation of K. variicola and K. quasipneumoniae from K. pneumoniae will help the adequate classification of Klebsiella species. This work contributes to recognizing the extensive presence of K. variicola and K. quasipneumoniae in diverse sites and samples. The correct identification of K. variicola in the hospital setting could contribute to a more appropriate medical treatment.17 Notably, some K. variicola18,19 and K. quasipneumoniae,20 respectively, have acquired carbapenem resistance and virulence factors.











 nueva página del texto (beta)
nueva página del texto (beta)