Introduction
Aedes aegypti is a major vector of infectious agents causing human disease at a global scale. The infectious agents transmitted by this mosquito include dengue, yellow fever, chikungunya, and Zika viruses. Currently, dengue is considered the most important viral disease transmitted by vector mosquitoes at a global scale. Recent estimations suggest that there are about 390 million dengue virus infections per year, a quarter of which become symptomatic.1 The dengue virus is transmitted between humans by the bite of mosquitoes from the Aedes genus, mainly A. aegypti.2 Most prevention strategies of dengue virus transmission are currently based on vector control. However, the impact of such strategies has been difficult to characterize, and it has not prevented the spread of mosquito populations.3 Therefore, the outbreaks of this and other viral diseases, with various degrees of public health and economic impact, continue to arise in many countries. A better understanding of vector-virus interactions should contribute to generate more focused strategies to prevent and control viral transmission. Vector-virus interactions are critical for the transmission of these pathological agents to human populations. The midgut is a critical point of entry for the initiation of the infection process leading to viral replication inside mosquito tissues.4,5 Once a virus has overcome the potential entry and the midgut escape barriers, it is disseminated to other tissues, where it overcomes other barriers until it reaches the salivary gland, from where it can be transmitted to humans (figure 1).
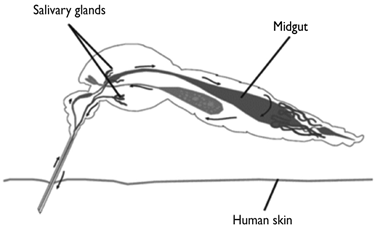
Black arrows show the trajectory of dengue virus replication and its dissemination within the vector mosquito A. aegypti. Mosquitoes acquire viruses by feeding on the blood of infected humans, after which their midgut epithelium cells can become infected. Viral replication inside the midgut epithelial cells shed virions that can reach the hemocele, other mosquito tissues, and eventually infect the salivary glands. Replication and shedding of the dengue virus in the salivary glands enables mosquitoes to transmit the virus by biting susceptible individuals.
Figure 1 Diagram of A. aegypti infection by dengue virus
To better understand the vector-virus interactions, some studies have been carried out to characterize the transcriptional profile of the mosquito midgut throughout the viral infection. The dengue virus infection of the cells of the mosquito midgut induces an extensive change on their genome-wide transcriptional profile.6,7,8 However, the precise molecular mechanisms that lead to a modified transcriptional profile of the midgut cells upon viral infection are not yet fully understood. Identifying the molecules and the mechanisms involved in the transcriptional regulation during a viral infection is essential to fully understand the vector-virus interactions that allow the virus to successfully replicate inside the mosquito cells. The participation of the cis-regulatory sequences is a key element in the transcriptional regulation, since these sequences serve as integrative nodes of gene regulatory networks. They function as docking sites for the formation of multiprotein complexes that ultimately recruit and direct RNA polymerase II to initiate transcription. Cis-regulatory sequences have a modular nature and multiple transcription factor binding sites. Identifying and characterizing the sequence of the cis-regulatory sites allows one to predict which specific transcription factors (TF) may participate in the gene regulatory networks. The binding of the transcription factors to form multiprotein complexes in cis-regulatory sequences requires the displacement of nucleosomes. In the case of many cis-regulatory sites, this displacement occurs during development, thus generating a specific open chromatin landscape for each cell type, according to the set of active genes that define its function. However, some cis-regulatory sequences are activated by chromatin remodeling only when the cell receives an external stimulus. Although cis-regulatory sequences have been identified in A. aegypti embryos,9 little is known about the cis-regulatory sequences that participate in the expression of genes during the viral infection of the mosquito midgut cells. Therefore, we set out to gain insight into the potential mechanisms involved in the viral infection-induced up-regulation of the mosquito genes.
The mosquito’s innate immunity system becomes activated upon invasion by a diversity of microbial agents, including viruses. Immunity genes have been identified in mosquito genomes,10 and the transcriptional activity of some of them is modified upon viral infection.11,12 More specific studies have revealed the participation of the Jak-STAT immunity pathway and its putative target genes in the mosquito’s anti-dengue defense.13 However, there are also genes whose function remains unknown, despite the fact that they undergo specific changes in expression during the viral infection. Such is the case of the AAEL006536 gene that becomes and remains up-regulated during the course of the dengue virus infection in A. aegypti midgut cells. This gene shows the highest increase, and it remains highly expressed seven days after feeding the mosquitoes with a DENV blood meal.7 It is also upregulated in the wMel Wolbachia infected A. aegypti strain.14 We used this gene as a model to investigate the potential mechanisms and molecules involved in the dengue virus-induced transcriptional up-regulation. Since open chromatin is a hallmark of the cis-regulatory function, we carried out an open chromatin profiling of the proximal upstream region of this gene in the dengue virus-infected mosquitoes. Here we report the following events: i) chromatin remodeling of the AAEL006536 gene proximal upstream region upon infection with DENV2; ii) identification of two cis-regulatory sites: one overlapping the transcriptional start site (TSS) and one upstream of the TSS, potentially corresponding to the promoter, and iii) sequence composition of the site upstream of the TSS, which indicated that this gene could be regulated by REL-, STAT- and C/EBP-type TFs. These data suggest that the AAEL006536 gene could be directly regulated by the mosquito’s immunity pathways and that it could be involved in the mosquito’s innate immunity.
Materials and methods
Mosquitoes. Female mosquitoes from the A. aegypti Rockefeller strain were used for blood feedings and dengue virus infections. Mosquitoes were obtained from the insectary at the National Institute of Public Health (Instituto Nacional de Salud Pública, Cuernavaca, México). Three to five-day-old females were used in this study. Mosquitoes were reared under a photoperiod cycle of 12 h light:12 h dark, at 28°C and 70 - 80% relative humidity. An antibiotic-antimycotic agent was added while they were fed with sugar (200 U/mL penicillin, 200 µg/mL streptomycin and 0.5 µg/mL amphotericin, Gibco BRL).
Dengue virus. Dengue virus serotype 2 (New Guinea C strain) was amplified by consecutive passes through Balb/c mice brains (2-3-day-old mice). Mice were sacrificed with CO2 vapor 5 to 6 days after viral inoculation, and their brains were stored at -70°C. The dengue virus was obtained from a brain homogenized in a phosphate buffered saline: fetal calf serum solution (1:1), at 30% w/v. The extract was clarified at 10 000 rpm for 30 min at 4°C and aliquots were stored at -70°C.
Dengue virus infection. Infection of mosquitoes with the dengue virus were carried out as previously described.15 The passage number history of the NGC strain used was C6/36 3, 4 SMB.
Dengue virus infection assay. RNA was extracted from the mosquito carcasses with TRIZOL reagent according to the manufacturer’s instructions. Complementary DNA synthesis was carried out with Promega Reverse Transcriptase for 60 min at 42°C according to the manufacturer’s instructions. The PCR reactions on cDNA template were carried out as previously described.15
Open chromatin assay. The formaldehyde-assisted identification of regulatory elements (FAIRE) technique was carried out as described16,17,18 on formaldehyde fixed pools of dissected midguts. Mosquitoes were dissected 8 days after feeding them. The midguts were fixed in formaldehyde-containing Drosophila Schneider’s medium, and the carcasses were frozen and stored to assess the rate of infection by RT-PCR.
qPCR. Overlapping amplicons were designed to cover approximately 2 Kb of the AAEL006536 gene proximal upstream region. PCR reactions were carried out with the SYBR Advantage kit (Clontech), following the manufacturer’s instructions. Amplicon enrichment was determined by the delta-delta Ct method of FAIRE vs control samples. The enrichment for all amplicons directed to the proximal upstream region was normalized using a coding region assumed to be associated with nucleosomes.
Sequence analysis. The MATCH software19 and the TRANSFAC database20 were used at the gene-regulation web server to search for putative transcription factor binding sites (http://www.gene-regulation.com/pub/programs.html.match).
Results
The FAIRE-qPCR analysis of the midguts of the blood-fed mosquitoes did not detect any open chromatin cis-regulatory sites in the AAEL006536 gene proximal upstream region. As figure 2(A) shows, most amplicons were depleted or only slightly enriched in these midgut FAIRE samples, which indicates that in non-infected midgut cells this proximal region is associated with nucleosomes in a closely packaged chromatin conformation with limited or no accessibility. This would be consistent with the absence of activated transcription of this gene in such a physiological state. In contrast to the previous condition, viral infection induces a clear change in the chromatin structure of the same proximal region, as shown in figure 2(B). FAIRE-qPCR analyses showed two distinct nucleosome-depleted cis-regulatory sites. One of these sites was overlapping the annotated transcriptional start site (TSS).
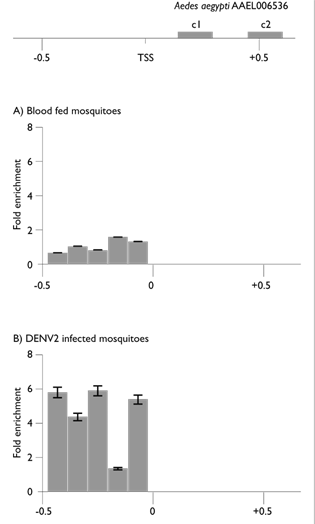
The top diagram depicts locus AAEL006536 (not to scale). Coding regions are depicted as black boxes (c1 and c2); TSS, transcriptional start site. Panels show the open chromatin profiles in (A) blood-fed mosquito midguts (8 days pbm) and (B) dengue virus-infected mosquito midguts (8 days pbm). The coordinates are in kilobases with respect to the TSS.
Figure 2 Open chromatin profile at locus AAEL006536
As it has been previously pointed out, the FAIRE assay for open chromatin profiling has the ability to detect active transcriptional start sites.18 The location and chromatin conformation of this FAIRE-enriched amplicon, over the TSS, is consistent with the transcriptional activation of this gene in dengue virus-infected mosquito midgut cells, as previously reported.7 A second open chromatin site was found upstream of the TSS. This open chromatin site was flanked by repetitive sequences at its upstream region. There was also a region of nearly 100 bp where all oligonucleotides designed did not result in an amplified fragment under any of the PCR conditions tested. Therefore, this region remained unexplored. This region may or may not be part of the detected upstream cis-regulatory site. Nevertheless, the open chromatin site, upstream of the TSS, spanned about 400 bp, considering the amplicons that became enriched. This region was in open chromatin conformation only in the dengue virus-infected mosquito midgut FAIRE samples. These results indicate that active chromatin remodeling is triggered by a dengue virus infection, generating a nucleosome-depleted region (NDR). The location of this cis-regulatory site, upstream of the TSS, suggests it could have a viral infection-induced promoter function in the transcription of this gene. If this were the case, this cis-regulatory site should contain binding sites for TFs. Therefore the sequence composition of this cis-regulatory site was analyzed by comparison to the Transfac database20 using the web server provided by Biobase.
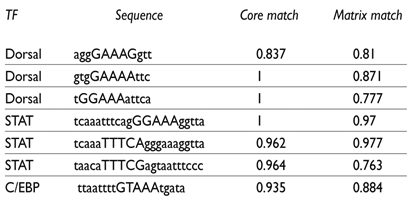
The sequence analysis of the upstream cis-regulatory site, which was activated by the chromatin remodeling during the dengue virus infection, identified a number of putative binding sites for immunity TFs. A comparison with the immunity TFBSs database confirmed the enrichment of immunity transcription factor binding sites, including STAT and NF-kappaB binding sites. On the other hand, a comparison with the insect database identified multiple binding sites for the Drosophila immunity TF Dorsal (NF-kappaB-type TF) and for the STAT family of TFs (table I). STAT transcription factors have been identified in Anopheles gambiae,21 and putative orthologues have been identified in A. aegypti and in the genomes of other mosquito species. The most frequent site found was the NF-kappaB-type TFBS, which is a member of the NF-kappaB family of TFs. Two TFs belonging to this family (REL1 and REL2), which participate in innate immunity, have been identified in A. gambiae.22,23 Therefore, the Dorsal type binding sites, found in the cis-regulatory sequence of the AAEL006536 gene, could be bound by a REL-type A. aegypti orthologue during the dengue virus infection. These findings suggest that the AAEL006536 gene could be regulated by immunity transcription factors responsive to the IMD and/or to the Toll immunity pathways. This would also suggest that this gene might be involved in A. aegypti’s innate immunity function. A C/EBP-type binding site was also identified in this cis-regulatory site. The analysis of an immunity gene promoter in A. gambiae has shown the participation of this type of TF in immunity transcriptional regulation.24 Table I shows the putative TFBS with the highest matching scores for the core of the sequence and the complete TFBS sequences.
Discussion
Here we have shown that the dengue virus infection induces chromatin remodeling at the proximal upstream region of the AAEL006536 gene in A. aegypti midgut cells. The displacement of nucleosomes in the cis-regulatory sequences is necessary to allow the binding of activating transcription factors. For some genes, this displacement is accomplished during development, when the cell-specific genome-wide open chromatin landscape is established, according to the cell’s function and phenotype.25 For other more tightly regulated genes, nucleosome displacement for cis-regulatory sequence activation may only be achieved upon stimulation, which leads to gene up-regulation. The infection-induced chromatin remodeling reported here results in the formation of two distinct open chromatin sites. One of them overlaps the TSS; it is consistent with an activated TSS, and it is detected only in dengue virus-infected mosquito midgut cells. An additional open chromatin site is formed upstream of the TSS. The sequence analysis of this proximal upstream cis-regulatory site revealed the presence of immunity transcription factor binding sites. Binding sites for STAT-type TFs suggest this gene could be a target of the Jak-STAT immunity pathway. Previous studies have shown that the Jak-STAT immunity pathway is involved in the antiviral response of mosquitoes,13 specifically the anti-dengue response. Putative target genes of this pathway, which participate in the mosquito’s anti-dengue defense, have been identified.26 The presence of REL-type TFBS suggests that this gene is a target of the Imd and/or the Toll immunity pathways.27 The finding of more than one class of immunity TF binding sites suggests that this cis-regulatory site may integrate signals that activate more than one innate immunity pathway.
The molecular model depicted in figure 3 summarizes the findings reported in this work. First, according to chromatin structure profiling in non-infected mosquitoes, there are no open chromatin sites in the proximal upstream region of the AAEL006536 gene. However, upon viral infection, the chromatin structure of this same region is remodeled, and two open chromatin sites are generated. The formation of these open chromatin sites implies the displacement of nucleosomes: at least one in the case of the TSS overlapping site and two to three in the case of the site upstream of the TSS. The analysis of the open chromatin site upstream of the TSS revealed the presence of STAT-, REL- and C/EBP-type TFBSs. These findings suggest that some or all of these transcription factors bind to this cis-regulatory site upon dengue virus infection. These findings also point to some form of cooperation (direct or indirect, through potential cofactors) between immunity TFs and chromatin remodeling factors (ChR F in figure 3).

This diagram shows the proposed chromatin conformation of the AAEL006536 gene’s proximal upstream region before and after a dengue virus infection in mosquito midguts. When midgut cells become infected with the dengue virus, nucleosomes are displaced generating an open chromatin site. This displacement may allow the binding of transcription factors and the formation of a multiprotein complex. This complex may then act as an active promoter leading to an increase in the transcription of this gene. TSS, transcriptional start site; REL, REL-type transcription factor; STAT, STAT-type transcription factor; ChR F, chromatin remodeling factor
Figure 3 Model of chromatin dynamics at the proximal upstream region of AAEL006536
The function of the AAEL006536 gene is currently unknown. No homologs of known function have been found, and it has not been classified as an immunity gene. However, viral infection up-regulates this gene, induces chromatin remodeling, and activates a cis-regulatory site located upstream of its annotated TSS. Furthermore, this upstream cis-regulatory site contains putative binding sites for REL- and STAT-type immunity transcription factors. These findings, together with the previously reported transcriptional up-regulation during viral infection, point to the possibility that this gene may have an immunity-related function. The best described immunity cis-regulatory site for A. gambiae is the Defensin 1 promoter that contains clustered REL-type binding sites and a C/EBP site.24 The presence of these two types of binding sites in the cis-regulatory sequence upstream of the AAEL006536 TSS supports its potential function as a promoter of an immunity gene. Findings reported in this work warrant further research into the regulatory function of this cis-regulatory site and the biological function of this gene.
In conclusion, the data presented here are consistent with the activation of this gene by chromatin remodeling at its upstream cis-regulatory sequences. Taken together, all these elements -up-regulation of this gene upon viral infection, chromatin remodeling, and the presence of immunity TF binding sites at the cis-regulatory sequence upstream of the TSS- suggest that this gene may play a role in A. aegypti’s anti-viral innate immunity function.











 nueva página del texto (beta)
nueva página del texto (beta)