Introduction
The taxonomy of the genus Klebsiella has been periodically revised. Currently, this genus includes K. pneumoniae subsp. pneumoniae, also known as KpI, and four novel species: Klebsiella quasipneumoniae, also known as KpII - with two subspecies: quasipneumoniae (KpII-A) and similipneumoniae (KpII-B); Klebsiella variicola (KpIII) and Klebsiella michiganensis.1 K. variicola and K. quasipneumoniae are sister species of K. pneumoniae. K. variicola has been isolated from plant tissues,2,3 fungal gardens of leaf-cutter ant colonies,4 cotton disease vectors (specifically of the insect, Nezara viridula),5 human6,7,8,9,10 and animal infections11 including bovine mastitis.12 Unlike K. variicola, K. quasipneumoniae has been described exclusively in hospital settings.13 A recent genomic comparative study with a large number of Klebsiella genomes showed that the nif operon was detected in all the genomes of K. variicola, in half of those of K. quasipneumoniae, but only in one strain of K. pneumoniae.13 Additionally, K. pneumoniae and K. variicola shared some virulence determinants that cause infections in humans.14,15
Some Klebsiella isolates have been misclassified16,17 and consequently the taxonomic descriptions of their genome sequences in NCBI repositories are inaccurate. Here we used correctly identified K. variicola and K. quasipneumoniae genomes for the comparison of virulence and plant colonization determinants of this genus of enterobacteria.
Materials and methods
K. variicola and K. quasipneumoniae genomes included in the study
A total of 41 Klebsiella genomes were included in the study, 31 of which corresponded to K. variicola, eight to K. quasipneumoniae and two to K. pneumoniae (table I). Of these genomes, 19 of K. variicola and six K. quasipneumoniae were originally misidentified.
Table I Origin and accession number data of K. variicola, K. quasipneumoniae and K. pneumoniae genomes included in the study

* The true bacterial specie was described previously by Chen and colleagues,16 and Martinez-Romero and colleagues.17
‡ Klebsiella quasipneumoniae subsp. quasipneumoniae
§ Klebsiella quasipneumoniae subsp. similipneumoniae
In silico analysis of virulence, plant-associated determinants and efflux pumps, regulators, heavy metal resistance and β-lactamases
A set of 138 proteins was selected from previous reports18,19 and the Pasteur Institute webpage (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html) for their putative roles in host interactions. Among these proteins, 84 were included for their involvement in virulence. Two efflux pump clusters, 11 efflux pump regulators, four heavy metal clusters, three β-lactamase family proteins (LEN-, SHV- and OKP-type), as well as 35 plant-association proteins that included nitrogen fixation enzymes (encoded by the nif operon with 20 genes), were also included. All genes encoding these proteins/enzymes were further searched in 41 Klebsiella spp. genomes by BLAST 20 (Genome BLASTp option with default values). A cluster analysis using the average linkage based on the Jacquard similarity coefficient and the respective dendrogram (constructed with UPGMA) were constructed using the DendroUPGMA program.20 In the case of the LEN-, SHV- and OKP-type families of β-lactamases, a phylogenic reconstruction was performed using the maximum-likelihood approach based on the JTT matrix-based model and 100 bootstrap replications (Mega v6.06).21 In addition, phylogenetic analyses were carried out on the amino acid sequences of FimV fimbrial proteins (KVR801v1_60088) and NifH nitrogenase (KVR801v1_120019) proteins.
PCR screening of fimV gene
Specific oligonucleotides for the fimV gene (fimV-F 5’-TTTGCGGATACTGACCAGGG-3’ and fimV-R 5’-GGTTACCACGGTCAGCGTAA-3’) were designed. Twenty-one K. variicola isolates from plants and humans were analyzed by PCR as previously described.8
In vitro assays of plant growth promotion
The mechanisms involved in plant growth promotion, such as phytohormone production (auxin [indole-3-acetic acid] and gibberellins), phosphate solubilization, siderophore production and lytic enzyme activities were evaluated in six K. variicola isolated from plants, and in 15 K. variicola and eight K. pneumoniae clinical isolates. The production of auxin was analyzed according to Khalid and collagues22 with Azotobacter vinelandii and Salmonella enteritidis as positive and negative controls, respectively. Phosphate solubilization was evaluated using the methodology described by Mehta and colleagues23 using Azotobacter vinelandii and E. coli DH5-α as bacterial controls. Siderophore production was evaluated using chrome azurol S according to Schwyn and Neilands24 with Pseudomonas fluorescens and Staphylococcus aureus as controls. Activities of lytic enzymes, such as lipases, proteases, esterases and amylases, were evaluated using the methodology described by Malleswari and Bagyanarayana25 using Bacillus subtilis and E. coli DH5-α as controls.
Ethical committee approval
This study is part of the K. variicola project that was revised and approved by the ethical commission from the Instituto Nacional de Salud Pública on the 21th of June 2011. The present study was carried out at the Instituto Nacional de Salud Pública and at the Centro de Ciencias Genómicas of UNAM, Cuernavaca, Morelos.
Results and discussion
Virulence-associated determinants in K. variicola and K. quasipneumoniae genomes
In silico screening of 84 virulence-determinant proteins in reported genomes of K. variicola, K. quasipneumoniae and K. pneumoniae showed a mosaic distribution in different isolates (table I). Most (>98%, except for FimV) of the virulence determinants that were selected here were originally described in K. pneumoniae NTUH-K2044. From the 84 virulence determinants included in the in silico analysis, 20 were found in at least one K. variicola genome and 11 in at least one K. quasipneumoniae genome. The proteins coded by the urease (UreA) and fimbriae gene cluster MrkABCDFHIJ were present in all the isolates of K. variicola and K. quasipneumoniae. The gene coding glucuronic acid transferase (WabG) was also present in all K. variicola and K. pneumoniae, but was absent in a single K. quasipneumoniae genome.
Siderophores, such as salmochelin (IroN), aerobactin (IucA and receptor IutA), KfuABC cluster, enterobactin (EntB) and yersiniabactin cluster YbtAESTX, were unequally distributed among Klebsiella. The most prevalent siderophores were enterobactin and KfuABC, in 93.5% (29/31) of K. variicola and in 100% (8/8) of the K. quasipneumoniae isolates. The aerobactin (IucA) was not identified, however the receptor of aerobactin (IutA) was present in 87% (27/31) and salmochelin (IroN) in 39% (12/31) of K. variicola isolates; however, in the K. quasipneumoniae genomes, both aerobactin receptor and salmochelin siderophore were present in 100% of the isolates. The siderophore yersiniabactin YbtAESTX cluster was identified in only one K. variicola isolate (MGH20) and in none of the K. quasipneumoniae genomes.
The genes encoding enzymes from the glycerate pathway (Gcl, GlxK), GlxR, and Hyi) were identified in K. variicola isolates (41.9 to 45.1%). The allantoinase cluster AllABCDRS genes and the YlbE-F and YbbW were present in few K. variicola genomes at 3.2, 6.4 and 3.2%, respectively. The GlxK-R and Hyi encoding genes were present only in K. quasipneumoniae genomes, in 75% of them. The genes for the two-component system KvgAS proteins were identified in 12.9% (4/31) of the K. variicola genomes and were absent from the K. quasipneumoniae genomes. The mucoviscosity-associated protein Wzy-K1 was identified in one K. variicola isolate (3.2%-1/31).
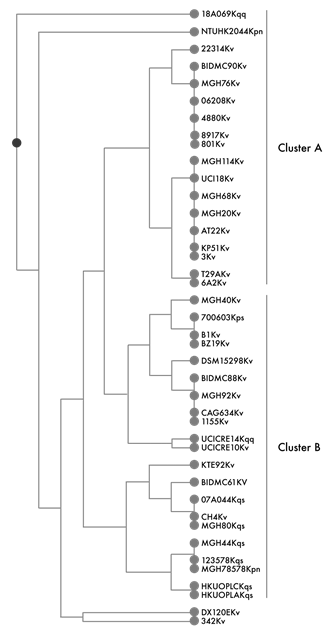
The Jacquard index analysis of the virulence-associated determinants that are shared between the K. variicola, K. quasipneumoniae and K. pneumoniae genomes is shown in figure 1. Three main clusters were obtained, two of which were closely related and could be distinguished only by a single siderophore difference. Cluster 1 grouped sixteen K. variicola and two K. quasipneumoniae subsp. similipneumoniae genomes that shared the siderophores IroN (11/18 genomes); receptor IutA, KfuABC and EntB (15/18 genomes); WabG, Uge, the fimbriae cluster MrkABCDFHIJ, FimV (15/18 genomes) and urease UreA. Cluster 2 includes ten K. variicola genomes and mostly contains the virulence-determinants proteins of cluster 1, except for IroN. Genes associated with all of the K. variicola genomes from cluster 2 were those encoding Glc, GlxKR and Hyi proteins that are involved in the glycerate pathway. This pathway together with allantoinase cluster genes are involved in allantoin metabolism as a nitrogen source.26 Cluster 3 consists of seven K. quasipneumoniae and one K. variicola genomes. All of the genomes of cluster 3 contain the Uge, WabG, IroN (except K. variicola KTE92 genome), IutA, KfuABC, EntB, UreA and MrkABCDFHIJ proteins. FimV protein was present in three (42.8%-3/7) of the K. quasipneumoniae genomes, corresponding to Klebsiella quasipneumoniae subsp. similipneumoniae (KpII-B).

Figure 1 Jacquard index of the virulence-determinant proteins of the K. variicola (Kv), K. quasipneumoniae (Kq) and K. pneumoniae (Kpn) genomes
K. pneumoniae NTUH-K2044 and K. pneumoniae MGH78578 each independently clustered together with K. pneumoniae BIDMC90 and K. variicola DSM15298, respectively (figure 1). The K. pneumoniae BIDMC90 genome shared with K. pneumoniae NTUH-K2044 fifteen virulence determinants. Finally, K. pneumoniae MGH78578 and K. variicola DSM15298 were different from the other clusters due to the absence of siderophore KfuABC, which was present in all of the K. variicola, K. quasipneumoniae and K. pneumoniae NTUH-K2044, and to the presence of the two-component system KvgAS, which was only present in the DX120E, MGH76 and UCICRE10 genomes (figure 1).
In silico analysis and prevalence of Fimbrial FimV protein
Previously a total of 114 unique proteins were identified in K. variicola genomes.8 Excluding transposons and tra-genes that are involved in horizontal transfer and selecting those that are involved in metabolism and cellular structure, 13 structural proteins were found to be unique for K. variicola, from which a fimbrial protein was chosen for further analysis (fimbria have been shown to mediate host interaction). The fimbrial FimV protein was identified in 90.3% (28/31) of the K. variicola (except for KTE92, B1 and BIDMC90) and in 37.5% (3/8) of K. quasipneumoniae but not in K. pneumoniae NTUH-K2044 and MGH78578 genomes. An additional BLASTp search in GenBank using a non-redundant protein sequence (nr-BLASTp protein-protein BLAST) identified the FimV protein in K. variicola genomes described in GenBank (except for KTE92, B1 and BIDMC90) and in the misclassified K. pneumoniae genomes that actually corresponded to K. variicola species (table I). The phylogenetic analysis of FimV proteins generally showed high amino acid conservation in K. variicola (>98%) (data not shown); however, in a few K. variicola strains, for example the wrongly named “K. pneumoniae 342”, a lower amino acid identity was observed (92 to 94%). Conversely, K. quasipneumoniae subsp. similipneumoniae, 07A044, 700603 and MGH44 FimV amino acid sequence had 91% similarity. FimV protein sequence was not found in bona fide K. pneumoniae; nevertheless it was identified in related Enterobacteriaceae such as Citrobacter koseri, Citrobacter freundii, Enterobacter, Salmonella enterica and Escherichia albertii, all with an amino acid similarity less than 53%. The FimV protein is also widely distributed in Escherichia coli and Shigella sonnei, with an amino acid identity of 33% to that from K. variicola.
The PCR screening of the fimV gene was carried out in a collection of twenty-one K. variicola isolates (see Materials and methods). The fimV gene was identified in 86.9% (20/23) of the K. variicola isolates. This prevalence is similar to that from in silico analysis (90.3%). This indicates that the fimV gene is not universally encoded in K. variicola genomes.
Plant-associated proteins in the K. variicola, K. quasipneumoniae and K. pneumoniae genomes
Plant-associated proteins were identified in K. variicola and K. quasipneumoniae isolates. Twenty-seven plant-associated determinants were found in at least one K. variicola or K. quasipneumoniae isolate. In general, 21 plant-associated determinants were contained in all of the K. variicola, K. quasipneumoniae and K. pneumoniae genomes that were included in the study. The NifJ-NifQ nitrogen fixation cluster was identified in all of the K. variicola genomes, in 62.5% (5/8) of K. quasipneumoniae genomes and was absent from the K. pneumoniae NTUH-K2044 and MGH78578 genomes. The NifH protein has high amino acid identity among all K. variicola genomes and in a phylogenetic analysis it is grouped in a cluster different from the corresponding sequence from the K. quasipneumoniae or other species. Holt and colleagues13 identified only one K. pneumoniae genome that contained the nifJ-nifQ operon. In this work, we identified another K. pneumoniae genome (from KPNIN29) that contains the nif operon in addition to that described by Holt and colleagues.13 Hazen and colleagues27 identified the absence of nifJ-nifQ gene cluster in K. pneumoniae NTUH-K2044, MGH-78578, 1162281, JH1, MS 92-3, 1191100241, ATCC13884 and KCTC-2242 genomes. Here we identified a conserved genetic context and gene synteny of nifJ-nifQ operon both in K. variicola and K. quasipneumoniae genomes (data not shown). Our results confirm that nitrogen fixation seems to be a characteristic trait of K. variicola.
Other differences identified in this work between the K. variicola, K. quasipneumoniae and K. pneumoniae genomes are genes encoding cellulases (CelK and BglH), catalases (KPK_4954 and KPK_2333) and hemagglutinins (HecA). While BglX cellulose is present in all K. variicola, K. quasipneumoniae and K. pneumoniae isolates, CelK and BglH were absent from K. pneumoniae NTUH-K2044 and MGH78578 and from 87.5% of the K. quasipneumoniae isolates.
The BglH enzyme, which has specificity towards 1,4-b glucosidic bonds and that most likely acts by hydrolyzing short cello-oligosaccharides, was present in 61.2% (19/31) of K. variicola isolates. The CelK gene encoding an enzyme for the decomposition of highly ordered forms of insoluble cellulose was present in K. variicola strains obtained from three different plants 342, T29A and 6A2 representing 10% (3/31) of the isolates. KPK_2233 (catalase) gene was present in 22.5% of the K. variicola genomes (7/31). KPK_4954 gene encoding a cyclic beta 1-2 glycan synthase possibly playing a role in osmotic adaptation was present in 16.1% of the K. variicola isolates (5/31).
HecA/B hemolysin/hemagglutinin secretion protein is involved in plant attachment in Erwinia chrysanthemis. A hecA gene mutant had reduced attachment, cell aggregate formation, and virulence in its host. HecA protein was present in 64.5% (20/31) of the K. variicola isolates. In K. quasipneumoniae, HecA protein is present only in HKUOPLA28 and HKUOPLC29 isolates obtained from giant panda feces.
The Jacquard index analysis showed two main clusters (A and B) that were grouped by the absence or presence of HecA, CelK or BglH protein (figure 2). Cluster A included the K. variicola and K. quasipneumoniae genomes that did not contain the HecA and CelK proteins. The K. quasipneumoniae 18A069 genome is the unique genome of this species that does not contain the YqeF (acetyl-CoA acetyltransferase), induce plant colonization and Ada (a regulatory protein of adaptive response) and DinF (DNA-damage-inducible protein F) putative stress response proteins.18
Efflux pump, regulators, heavy metal resistance and chromosomal β-lactamase proteins in K. variicola, K. quasipneumoniae and K. pneumonia
The OqxABR efflux pump gene clusters are present in all K. variicola and K. quasipneumoniae genomes examined. The AcrABR protein efflux was identified in 58.0% (18/31) and 100% (8/8), of K. variicola and K. quasipneumoniae. All of the protein regulators were identified (MarAR, SoxSR, RamAR, Rob, SdiA, Fis, EnvR, and RarA) in >98% and 100%, of the K. variicola and K. quasipneumoniae isolates. All efflux pumps and efflux pump regulator proteins were identified in the K. pneumoniae NTUH-K2044 and MGH78578 genomes.
The copper (PcoABCDERS), silver (SilCERS) and tellurium (TerABCDEWXYZ) protein clusters for heavy metal resistance were analyzed. The PcoABCDERS and SilCERS protein clusters were identified together in 22.5% (7/31) and 37.5% (3/8) of the K. variicola and K. quasipneumoniae genomes, respectively, while the K. variicola Bz19 genome contained only the PcoABCDERS protein cluster.
Tellurite resistance is conferred by the TerABCDEWXYZ protein cluster and seemingly is strongly associated with the hypervirulent clonal groups.30 Tellurite resistance is needed to colonize macrophages.31 The TerABCDEWXYZ proteins cluster was identified in 12.9% (4/31) and 12.5% (1/8), of K. variicola and K. quasipneumoniae, all of human origin. K. variicola 8917 isolate was described as hypermucoviscous.15
In the analysis of chromosomal β-lactamase, we found that all the K. pneumoniae genomes that were identified as K. variicola contained the LEN-type β-lactamases allele. The K. pneumoniae genomes that were correctly identified as K. quasipneumoniae contained the OKP-type alleles, while the K. pneumoniae NTUH-K2044 and MGH78578 genomes contained the SHV-type alleles. The chromosome-encoded β-lactamases corresponded to constitutive genes of these bacterial species (figure 3A). The OKP-A1 and OKP-B1 β-lactamases is characteristic of K. quasipneumoniae subsp. quasipneumoniae (KpII-A) and K. quasipneumoniae subsp. similipneumoniae (KpII-B), respectively.

Figure 3 Phylogenetic analysis of chromosomal b-lactamases. A) LEN-, SHV- and OKP-type b-lactamases that were identified in the K. variicola, K. quasipneumoniae and K. pneumoniae genomes. B) Chromosomal b-lactamase protein that was annotated in K. quasipneumoniae genomes and that corresponded tothe OKPfamily. HPLAand HPLC, respectively, corresponds toK. quasipneumoniae subsp. similipneumoniae HKUOPLAand HKUOPLC
β-lactamase genes were present in most of the K. variicola and K. quasipneumoniae genomes and were previously annotated as “Beta-lactamase or class A beta-lactamase”. In K. pneumoniae B1 and K. variicola, BZ19 the corresponding sequence was annotated as “Beta-lactamase TEM” and had an amino acid identity of 99 and 100%, respectively, with LEN-2 b-lactamase. In the Klebsiella sp. 1.1.55 genome, β-lactamase was annotated as “LEN family Class A β-lactamase” and had a 99% of amino acid identity with LEN-2. Figure 3B shows the phylogenetic analysis of K. quasipneumoniae chromosomal β-lactamase genes. The β-lactamases were annotated as class A beta-lactamase, or SHV-1 or TEM proteins. However, all these proteins corresponded to OKP-A1 and OKP-B1 β-lactamases, respectively, in subspecies of K. quasipneumoniae (KpII-A, KpII-B) (figure 3B). Chromosomal LEN- and OKP-β-lactamase proteins were not properly annotated and could have contributed to the bacterial species misclassification, if these chromosomal β-lactamase proteins are considered for Klebsiella classification.
In vitro assays of plant growth-promoting mechanisms in K. variicola and K. pneumonia
Indole acetic acid (an auxin) has the ability to regulate root growth, differentiation of vascular tissues, elongation, apical dominance, initiation of lateral roots and maturation.32 This molecule also functions as an important signaling molecule in the regulation of plant growth, expansion, cell differentiation and cell division regulation.33K. variicola and K. pneumoniae isolates were capable of producing indole acetic acid (table II).
Table II In vitro assays of plants growth-promoting mechanisms in K. variicola and K. pneumoniae isolates
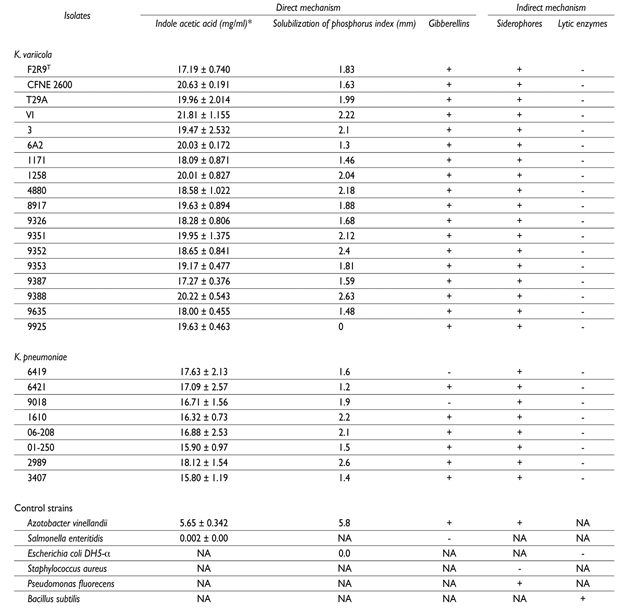
* The in vitro assay was carried by triplicated and the standard errors are shown
NA; not applied, +; positive, -; negative
Gibberellins (GA) are involved in seed germination, seedling emergence, stem and leaf growth, flower induction, vegetative regulation of reproductive dormancy buds and fruit growth.34,35 Almost all of the K. variicola and K. pneumoniae isolates (except for two K. pneumoniae isolates) were capable of producing gibberellins.
In addition to nitrogen, phosphorus is a nutrient that is required by plants.36 K. variicola and K. pneumoniae isolates were not able to solubilize phosphate (table II), which is a common characteristic among plant associated bacteria. Klebsiella genus, unlike other plant growth promoting bacteria, did not produce chitinases or proteases that may help fighting pathogenic fungi of plants (table II). Siderophores are synthesized mainly by Gram-negative bacteria, fungi, yeast and some plants (phytosiderophores) and act as specific chelating agents. They are soluble in aqueous solutions at neutral pH and are considered as secondary metabolites.37 Siderophores may be important in the process of colonizing plant and human tissues.38 Different siderophores were identified in silico in the genome of K. variicola, K. quasipneumoniae and K. pneumoniae. In agreement with these findings, the in vitro siderophore assay was positive for all of the K. variicola and K. pneumoniae isolates that were tested.
The name K. variicola means from various places and novel reports confirmed the adequacy of its name. This species has now been identified in diverse plants, such as bitter gourd and chili, as well as in insects such as Nezara viridula. It is also significant that isolates have been obtained from different human samples, such as urine, pus and bile.17
K. variicola isolates were additionally obtained from the fecal microbiota of two cohorts of Malawian infants/children, but their genomes could not be included in this work because they are fragmented into a high number of contigs. Of note, the K. variicola strain has been identified in giant panda feces,28 and corresponds to other example of misclassification and corresponds to K. quasipneumoniae.17 The first OXA-181 (carbapenemase)-producing K. variicola isolate was identified (by rpoB gene analysis) in fresh vegetables that were commercialized between different continents (Asia-Europe).39 This work showed the fist carbapenem-resistance K. variicola strain and may suggest possible emergence paths of resistance in Enterobacteriaceae.
Gene flow may occur among different related Klebsiella species, mediated by plasmid transfer. The plasmid of K. variicola pBz19 showed high identity to pl9 (both associated with IncN incompatibility) from K. pneumoniae isolate.14 K. variicola DX120E contains pKV1 and pKV2 plasmids. The pKV1 plasmid is very similar to the pKp5-1 plasmid that was identified in K. pneumoniae KP5-1.19 Plasmid pKV2 is most similar to plasmid pKOXM1C from K. oxytoca strain M1, suggesting plasmid exchange between these bacterial species. Additional analyses of plasmids are required specially in K. variicola from different niches to further understand their role in host interaction.
Conclusions
This work provides a new molecular framework for distinguishing different organisms of the Klebsiella genus. The in silico analyses showed that the genomes of K. variicola and K. quasipneumoniae shared a set of virulence-associated genes encoding mainly siderophores (aerobactin and enterobactin KfuABC cluster), urea metabolism, lipopolysaccharide biosynthesis enzymes (WabG and Uge), and fimbriae genes encoding the MrkABCDFHIJ gene cluster. The in vitro analysis indicated the presence of different siderophores in distinct isolates and species. Siderophores may have redundant functions given that they all are involved in bacterial iron acquisition. The number of siderophores in a single cell and their type however, may be important to regulate the flow of iron incorporation into the bacterial cell, a process that is linked to invasion of host tissues and thus virulence.
FimV, on the other hand, could be considered a common character in K. variicola strains distinguishing them from those of K. pneumoniae; however, not all K. variicola genomes contained this gene, as shown by both in silico and PCR assays. To explain the mosaic distribution of putative host-interaction genes in Klebsiella genomes, we envisage the possibility that Klebsiella cells may acquire virulence genes by horizontal transfer from closely related bacteria. However, this possibility is tempered by the alternative process of gene loss in bacterial evolution, which could be rather significant in the conformation of Klebsiella genomes as has been shown in other bacteria.40 K. variicola and K. quasipneumoniae strains with different genetic repertoires have the capacity to adapt to plant and clinical settings. Previously, we suggested that a different epidemiological dynamics occurred with K. variicola in comparison to K. pneumoniae with K. variicola coming from the environment,41 we further refine here that the environment refers most probably to the plant environment. Plants can be a reservoir for K. variicola isolates that may opportunistically infect humans or animals. K. variicola infection in humans thus seems to be a case of “phytonosis”, a term we suggest for symbiotic bacteria plant-borne, parallel to the term zoonosis bacteria pathogen animal-borne.











 nueva página del texto (beta)
nueva página del texto (beta)