Introduction
Inborn errors of metabolism (IEM) represent a group of about 500 rare genetic diseases, closely associated with intellectual developmental disorders (IDD), where the diversity of metabolic pathways involved explains the difficulties in making an accurate and early diagnosis.1,2 Basic biochemical procedures, including quantification of amino acids (AA) and acylcarnitines (AC), should be systematically performed whenever an IDD is suspected,3 to identify treatable congenital metabolic disorders (table I). Routine metabolic screening is especially important in countries like Mexico, where newborn screening (NBS) programs are still limited to only a few diseases4,5 and where IDD affect close to 2 million people under 18 years of age.6
IEM are a complex group of monogenic disorders leading to the accumulation of toxic compounds, cellular energy deficiency, or a lack of the substrates necessary for important biochemical processes. Although individually rare, collectively IEM represent the causes of devastating disturbances of the developing nervous system,7 including brain formation abnormalities and mild to severe mental disability.8,9 Opportune detection of IEM is essential because specific treatments may be available, metabolic decompensation could be avoided, and accurate genetic counseling can be provided, thereby offering the possibility of preventing the effects of IEM on brain development and function or reverting to some extent the consequences on mental health.
Table I IEM associated with IDD that could be detected through tandem-mass spectrometry metabolic screening
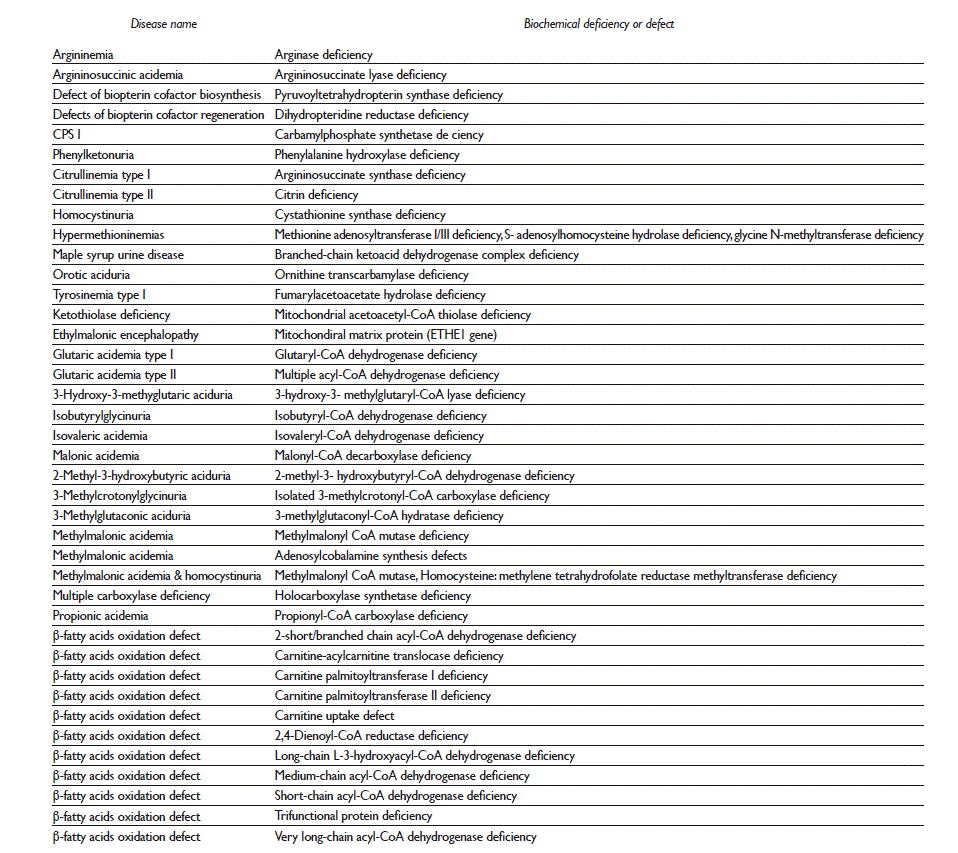
Metabolic screening comprises biochemical testing of blood, urine, or cerebrospinal fluid samples, to be used in the diagnosis of an IEM.10 In developed countries, the detection of IEM has been focused on newborns, so as to obtain the earliest diagnosis possible and receive prompt treatment. However, some studies highlight the importance of IEM detection in adults with IDD.11 NBS was first applied to massive detection of phenylketonuria, an IEM of amino acids. Initially, screening was done using a simple bacterial inhibition assay, but over time, technological advances have enabled the detection of many other metabolic disorders.12 Tandem mass spectrometry (MS/MS) is a powerful analytical tool that can be applied to metabolic screening both in neonates and in people of other ages. MS/MS methodology analyzes biological samples for both amino acids and acylcarnitines, among other metabolites.13 In a systematic literature review update in 2013, 89 treatable IEM that cause IDD have been identified, many of which can be detected by MS/MS7 (table I). Worldwide, there are important variations in the number and type of disorders detected through NBS. Congenital hypothyroidism is the most frequently screened disease, but mandatory screening for IEM only exists in developed countries.4 In Mexico, the NBS disease panel has also varied.14 Currently, the Ministry of Health only includes five diseases in the screening program: congenital hypothyroidism, congenital adrenal hyperplasia, cystic fibrosis, phenylketonuria, and galactosemia. Therefore, the number of IEM screened remains very low, which results in the high likelihood to find undiagnosed patients between the IDD Mexican population.
Recently, metabolomics has been used for the study of pediatric neurologic and psychiatric conditions such as Down syndrome,15 schizophrenia,16 and autism.17 Metabolomics could be used to identify unreported metabolic alterations associated with IDD. Metabolomics refers to the comprehensive measurement of small molecules, typically <1 500 daltons (e.g., sugars, amino acids, organic acids, nucleotides, acylcarnitines, and lipids), called metabolites, which are present in biological samples. Metabolomics is a potent tool for the study of human metabolism in health and disease.18 Comparative statistical analysis can reveal perturbations of metabolite levels in disease conditions and thus has the potential to identify novel biomarkers for diagnosis, prognosis, and treatment response. Metabolomics is complementary to genomics, transcriptomics, and proteomics.19 Metabolites represent the end products of the genome and proteome and can therefore be helpful in providing a holistic physiologic phenotype of a system or metabolic pathway.20 Metabolome profiling can be useful in disease heterogeneity for evaluating the underlying biological state of individuals through assessment of metabolite levels, thus providing a better understanding of disease mechanisms.
To identify novel metabolic diseases associated with IDD of unknown etiology and comorbidities, the objective of this study is to carry out an extensive metabolic analysis, including standard metabolic screening and metabolomics analysis, as part of the Intellectual Developmental Disorders Mexico Study (IDD Mexico Study).
Materials and methods
This methodology paper is part of the IDD Mexico Study, which has been reviewed and approved by the Ethics in Research Committee of the National Institute of Public Health, with number CI 1 456. The diagnostic algorithm shown in figure 1 will be followed to identify individuals with IDD of unknown etiology and comorbidities so as to conduct the metabolic screening and metabolomics analysis described herein, as well as the complementary genomics characterization previously presented.6
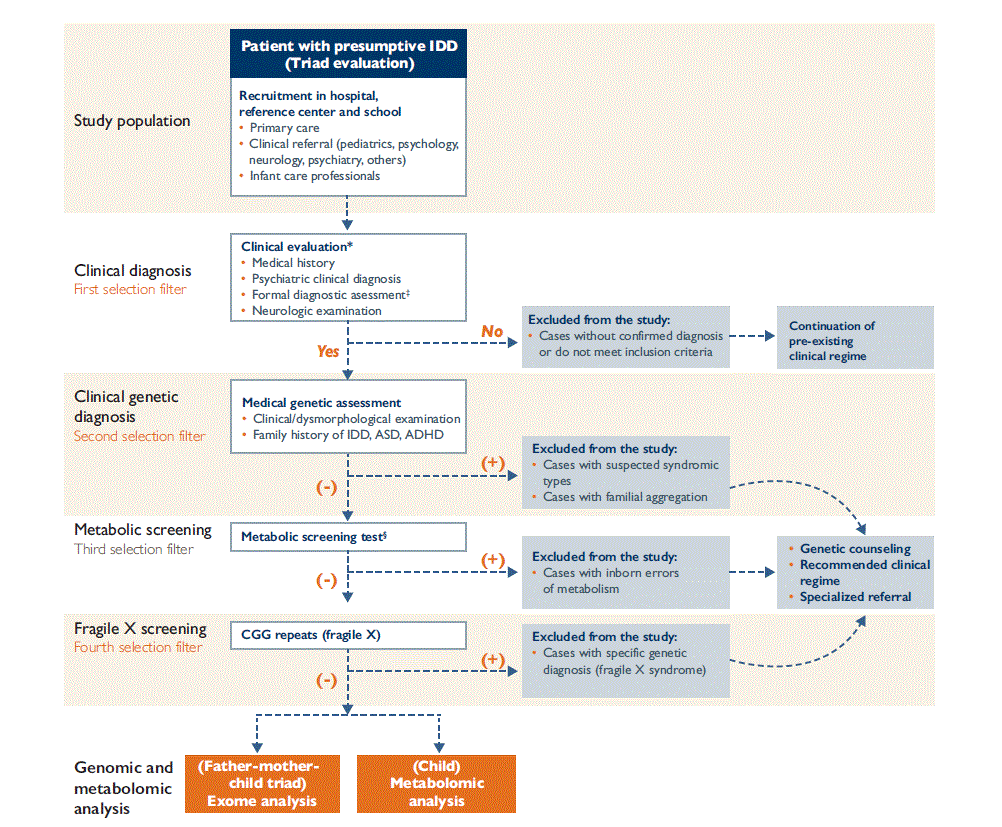
* Recruiting and evaluation of triads with children and adolescents 6-15 years of age will be carried out at the Dr. Juan N. Navarro Children´s Psychiatric Hospital (HPIDJNN) in Mexico City and recruitment and evaluation of triads of people over 18 years old will be done at the Integral training and development center (CADI A. C.) in Mexico
‡ Formal diagnostic evaluation will be done through application of tools described in the section on clinical diagnosis
§ Metabolic screening will be done at National Institute of Pediatrics (INP) in Mexico City. Children and adolescents with IDD or autism spectrum disorders will be evaluated, to exclude cases with inborn metabolism errors.
IDD: Intellectual Developmental Disorder, ADHD: Attention Deficit Hyperactivity Disorder, ASD: Autism Spectrum Disorder.
Algorithm modified from reference 6
Figure 1 Diagnostic algorithm for the evaluation of subjects with IDD of unknown etiology
Metabolic screening test
After clinical and clinical genetic diagnoses (first and second selection filters previously discussed by Lazcano and colleagues),6 metabolic screening (third selection filter) will be done at the Laboratory of Inborn Errors of Metabolism and Screening of the National Institute of Pediatrics, as follows:
Quantification of AA, AC, and succinylacetone (SA). Dried blood spots (DBS) will be analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS), using a Quattro micro API tandem MS with a commercial kit (NeoBase Non-derivatized MSMS Kit; PerkinElmer, Waltham, MA, USA). Quantification of the metabolites is achieved using appropriate internal standards as reference. This method has been proved to be in concordance with “Guidance for Industry-Bioanalytical Methods Validation”, which implies proof of reproducibility within a given error range.21 In cases of abnormal screening results, 4-5 drops of dry blood sample will be obtained for a second confirmatory quantification of AA, AC and SA. In addition, a 5-ml urine sample will be collected, to perform organic acid analysis by gas chromatography mass spectrometry (GC/MS) on an Agilent 6890N GC coupled to an MSD 5973 MS (Agilent Technologies, Santa Clara, CA, USA), as previously described. 5
Thyrotropin (TSH) determination. A 3-mm punch of each DBS sample will be used for TSH quantification with a commercial kit (hTSH FEIA PLUS) and a fluorometric equipment (Fluoroskan Ascent), both from Ani Labsystems Ltd. (Vantaa, Finland).
IEM associated with IDD that will be ruled out using this approach are shown in table I; additionally, congenital hypothyroidism will also be detected. Patients with an IEM will be excluded from the study and clinical and genetic counseling will be provided.
Metabolomics analyses
As shown in figure 1, offspring (children, adolescents, and young adults) with normal results for metabolic screening and negative results for Fragile X screening (the fourth selection filter discussed by Lazcano and colleagues)6 will undergo metabolomics analyses of DBS samples with LC-ESI-MS/MS, to identify metabolic profiles associated with pure and comorbid diagnostic of IDD of unknown etiology (see stratification previously described for genomic characterization).6 Statistical analysis will be performed using MetaboAnalyst.22 Partial least squares discriminant analysis (PLSDA) and variable importance in projection (VIP) scores will be used.
Discussion
In the metabolic screening to be carried out as part of the IDD Mexico Study, close to 50 congenital metabolic diseases will be analyzed, and patients identified with one of these disorders will receive appropriate medical attention. Furthermore, the study will yield information about the epidemiology and etiology of IDD, which in conjunction with other findings about children’s health,23 will support decision making for health services in Mexico.
The increased use of “omics” technologies offers the perspective of the molecular basis, to construct and understand the origin of pathologies and address them more efficiently. Metabolomics is a new technology based on the biochemical characterization of metabolites related to genetic and environmental changes.24,25 Metabolomics knowledge and characterization could complement the genomic approach of the IDD Mexico Study, for the first time providing metabolic insights for patients with IDD in Mexico.











 nueva página del texto (beta)
nueva página del texto (beta)


