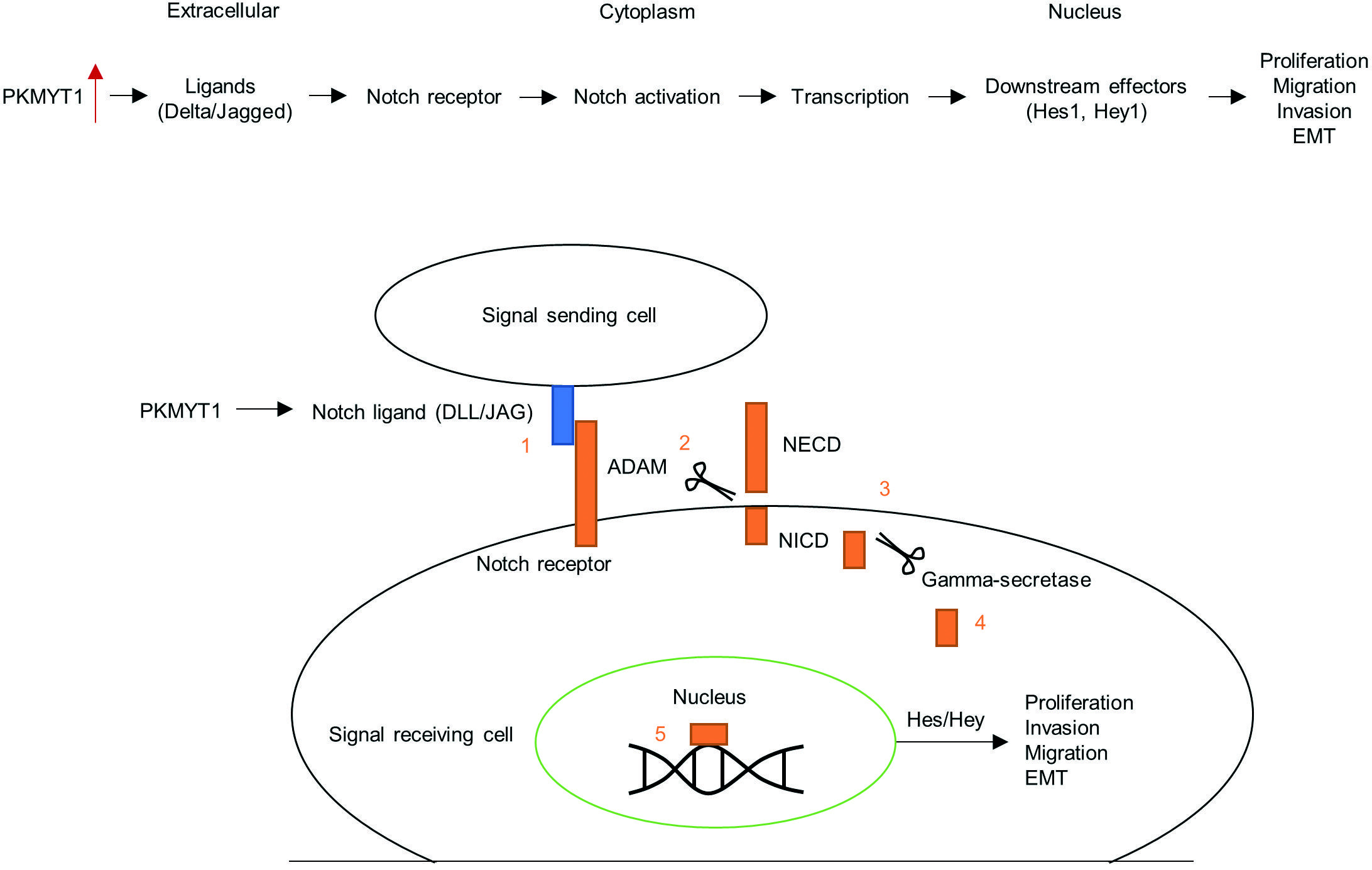
INTRODUCTION
Breast cancer (BC) is the most frequently diagnosed malignancy among women1. The most representative subtypes of BC are luminal A, luminal B, HER2 positive, and basal-like breast cancer. Luminal A, the most frequent BC subtype, is characterized by estrogen receptor (ER) and progesterone receptor (PR) positivity. Luminal B, like luminal A, is ER and PR positive, but it is differentiated by its high expression of proliferation markers, specifically Ki67. HER2-positive BC is classified by the overexpression of the tyrosine kinase family HER2 receptor. Finally, basal-like BC is classified as triple-negative (negative for PR, ER, and HER2 receptors)2. In 2022, there were approximately 429,105 and 259,827 new cases of female BC and almost 124,002 and 44,094 deaths from BC in China and the USA, respectively3. Although the patients survival outcomes have been greatly improved by currently available treatment methods4,5, local recurrence and distant metastasis remain the major cause of BC-related death6,7. Current adjuvant therapy for BC can also eliminate tumor cells that have spread to distant sites at the time of diagnosis and significantly improve the 10-year survival rate of women. However, approximately 40% of women who have already received adjuvant therapy would develop post-operative metastasis and eventually die of metastatic BC8. Moreover, current prognosis is mainly based on clinicopathological parameters such as lymph node status, tumor size, distant metastasis, histological grade, and tumor-node metastasis (TNM) stage. These are powerful prognostic indicators, but these may be only rough measures of the biological behavior of a tumor, and some of these parameters might be influenced by the subjectivity of the pathologist and limited in their prognostic value9. Triple-negative breast cancer (TNBC), which accounts for 15-20% of incident breast cancers, is the only BC subtype that lacks targeted treatments, and the aggressive biological and clinical behavior of TNBC translates into more frequent and earlier relapses than other subtypes of BC10,11. Therefore, further probing into therapeutic targets and prognosis biomarkers is beneficial for a higher survival rate of TNBC patients.
Distant metastasis is the leading cause of death in BC patients, especially in patients with TNBC12. Epithelial-to-mesenchymal transition (EMT) is an evolutionally conserved, reversible and dynamic cellular process, in which epithelial cells gradually lose cell-cell adhesion, undergo extensive cytoskeleton reorganization, change their cellular morphology, and become migratory and invasive mesenchymal cells13. TNBC cells that undergo an oncogenic EMT process facilitate the metastatic abilities of neighboring epithelial tumor cells14. This event is featured by N-cadherin acquisition and E-cadherin loss, and a group of transcription factors have been demonstrated to be capable of orchestrating EMT in cancer progress, including ZEB1 and Snail15. Thus, EMT inhibitors are recognized as potential targets to inhibit cancer progression.
Mammalian Notch signaling is initiated by receptor-ligand interactions between neighboring cells. Notch receptor activation results in the translocation of its intracellular domain into the nucleus to induce the expression of downstream target genes16,17. The canonical pathway involves translocation of the activated Notch cytoplasmic domain to the nucleus. Hes1 and Hey1 are among the key canonical Notch effector gene products18. p21/WAF1 protein is known as cyclin-dependent kinase inhibitor 1 and is able to bind to and inhibit the activity of cyclin-CDK2 complexes, thus regulating the G1 phase progression of the cell cycle. Activation of Notch1 signaling can promote p21 expression in TNBC cells19. Accumulating evidence has indicated that the Notch signaling plays an important role in the progression of several cancer, including TNBC20-22. Thus, identifying regulators of Notch degradation will provide a therapeutic strategy for TNBC treatment.
Protein kinase, membrane-associated tyrosine/threonine 1 (PKMYT1) is located at 16p13.323, and its level is found to be upregulated in BC and high PKMYT1 expression predicts poor prognosis in BC24. In addition, PKMYT1 expression in BC is associated with clinicopathologic features, and PKMYT1 downregulation can inhibit the proliferation, migration and invasion and promote the apoptosis of TNBC cells25. In addition, PKMYT1 serves as an oncogene in ovarian cancer26, gastric cancer27,28, prostate cancer29, lung adenocarcinoma30, and glioblastoma31. Based on these findings, we identified PKMYT1 as a prognostic biomarker in BC progression and explored the underlying mechanisms by which PKMYT1 affects the development of BC.
Consequently, this study was designed to investigate the related mechanisms of PKMYT1 in the development of TNBC. PKMYT1 upregulation has been found to activate Notch signaling in lung cancer32. We hypothesized that PKMYT1 might be a regulator of Notch signaling in TNBC progression. Thus, we conducted a series of experiments to verify the hypothesis. This study might provide a novel modulatory mechanism for TNBC.
METHODS
Patients and tissue samples
BC specimens were obtained from Wuhan NO.1 Hospital. Fifty pairs of surgically resected samples (tumor and adjacent non-tumor tissue samples) were collected from patients with a newly diagnosed BC who had received no therapy before sample collection. Collected samples were frozen in liquid nitrogen immediately and stored at −80ºC. All pathological documents, including patients' age, tumor size, lymph node status, histological grade, pathological tumor-node-metastasis (pTNM) stage, recurrent/metastasis, and subtypes, were carefully obtained and recorded in Table 1. Ethical approval was obtained from Clinical Research Ethics Committee of Wuhan NO.1 Hospital, and the written informed consents to use their tissues for scientific research were all signed by patients.
Table 1. Correlation between PKMYT1 and clinicopathological factors of patients with breast cancer
| Characteristics | n | PKMYT1 expression | p-value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Age | ||||
| < 50 | 27 | 15 (56) | 12 (44) | 0.3946 |
| ≥ 50 | 23 | 10 (43) | 13 (57) | |
| Tumor size (cm) | ||||
| < 2 | 26 | 11 (42) | 15 (58) | 0.2575 |
| ≥ 2 | 24 | 14 (58) | 10 (42) | |
| Lymph node metastasis | ||||
| Negative | 26 | 8 (31) | 18 (69) | 0.0046 |
| Positive | 24 | 17 (71) | 7 (29) | |
| Histological grade | ||||
| I | 19 | 4 (21) | 15 (79) | 0.0004 |
| II | 15 | 7 (47) | 8 (53) | |
| III-IV | 16 | 14 (88) | 2 (12) | |
| pTNM stage | ||||
| I | 17 | 3 (18) | 14 (82) | 0.0003 |
| II | 14 | 6 (43) | 8 (57) | |
| III-IV | 19 | 16 (84) | 3 (16) | |
| Recurrence/metastasis | ||||
| Yes | 23 | 18 (78) | 5 (22) | 0.0002 |
| No | 27 | 7 (26) | 20 (74) | |
| Subtypes | ||||
| Luminal | 14 | 8 (57) | 6 (43) | 0.1666 |
| HER2 positive | 14 | 4 (29) | 10 (71) | |
| Triple negative | 22 | 13 (59) | 9 (41) | |
All subjects gave their informed consent for inclusion before they participated in the study. The human tumor tissues used in this study were approved by the Clinical Research Ethics Committee of Wuhan NO.1 Hospital. All animal experiments and procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NIH) and approved by the Animal Care Ethics Committee of Wuhan NO.1 Hospital. All methods were performed in accordance with the relevant guidelines and regulations.
Culture of breast cancer cells
Human BC cell lines (MCF7, T47D, MDA-MB-436, and MDA-MB-231) and breast epithelial cell line (MCF10A) were acquired from Dalian Meilun Biotechnology (Liaoning, China) and incubated in Dulbecco's modified Eagle's medium (DMEM) (Solarbio, Beijing, China) containing 10% fetal bovine serum (FBS), 100 U/mL of penicillin and 100 pg/mL of streptomycin in a humidified incubator containing 5% CO2 at 37ºC. Cells were split 2 or 3 times weekly at a ratio between 1:2 and 1:4. MCF7 and T47D cell lines are estrogen receptor (ER)-positive, while MDA-MB-436 and MDA-MB-231 cell lines are triple-negative.
Treatment of breast cancer cells
To detect the effects of PKMYT1 and Notch on the malignant phenotypes of TNBC cells, we effectively downregulated PKMYT1 and Notch expression in TNBC cells. MDA-MB-436 and MDA-MB-231 cells were seeded into 6-well plates (2×105 cells/well) and allowed to grow for 24 h before being washed with phosphate buffered saline (PBS). When reaching 80% confluence, the cells were harvested and pelleted by centrifugation at 1300 rpm for 5 min. Then, the medium was aspirated, and the cells were resuspended in Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) before being transfected with 1 μg short hairpin (sh)-RNA against PKMYT1 (sh-PKMYT1#1/2) or 1 μg negative control shRNAs (sh-NC) obtained from GenePharma (Shanghai, China). Jagged 1 (MedChemExpress, Shanghai, China), a Notch activator, was added into MDA-MB-436 and MDA-MB-231 cells after intervening PKMYT1 expression. The cells were collected for further studies after 48 h of treatment. Transfection efficiency was validated by RT-qPCR and Western blotting.
Real-time quantitative polymerase chain reaction (RT-qPCR)
The mRNA levels of PKMYT1, Notch1, Notch2, and Notch3, were measured by RT-qPCR. Total RNA was separated from frozen samples or harvested cells using TRIzol reagent (Sigma-Aldrich, St-Louis, MO, USA), and cDNA was synthesized through reverse transcription of 1 μg total RNA through a Roche Transcriptor First Strand cDNA Synthesis kit (Solelybio, Shanghai, China). Subsequently, RT-qPCR was conducted using the SYBR Green qPCR assay kit (Solarbio) on an Applied Biosystems 7500 system (Applied Biosystems, Foster City, CA, USA). GAPDH was used as an internal control. The cycling protocol was 95°C for 30 s (initial denaturation), followed by 40 denaturation cycles at 95°C for 3 s, and finally annealing and extension at 60°C for 30 s. Relative mRNA expression was quantified using the 2−ΔΔCt method33. The primer sequences used in this study are listed in Table 2.
Table 2. Sequences of primers used for reverse transcription-quantitative PCR
| Genes | Sequence (5'→3') |
|---|---|
| PKMYT1 forward | CATGGCTCCTACGGAGAGGT |
| PKMYT1 reverse | ACATGGAACGCTTTACCGCAT |
| Notch1 forward | GAGGCGTGGCAGACTATGC |
| Notch1 reverse | CTTGTACTCCGTCAGCGTGA |
| Notch2 forward | CAACCGCAATGGAGGCTATG |
| Notch2 reverse | GCGAAGGCACAATCATCAATGTT |
| Notch3 forward | TGGCGACCTCACTTACGACT |
| Notch3 reverse | CACTGGCAGTTATAGGTGTTGAC |
| GAPDH forward | GGAGCGAGATCCCTCCAAAAT |
| GAPDH reverse | GGCTGTTGTCATACTTCTCATGG |
Western blotting
The protein levels of PKMYT1, E-cadherin, N-cadherin, Snail, ZEB1, Notch1, p21, Hes1, Hey1, Notch2, and Notch3 were evaluated by Western blotting. Total protein was extracted from TNBC cells and normal breast epithelial cells using ice-cold radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich) containing 1 mmol/L phenylmethylsulfonyl fluoride and protease inhibitor cocktail (1:100 dilution; Beyotime, Shanghai, China). Samples were centrifuged at 4°C for 15 min at 14,000 rpm, and supernatants were collected, followed by quantification of protein content using a BCA Protein Assay Kit (Innochem, Beijing, China). Thereafter, proteins (40 μg) were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted on a polyvinylidene difluoride membrane. After blocking with 5% skimmed milk for 2 h, the membrane was incubated overnight with primary antibodies against N-cadherin (ab98952, 1:1500; Abcam, Shanghai, China), PKMYT1 (ab200387, 1:500; Abcam), GAPDH (ab125247, 1:6000; Abcam), E-cadherin (ab1416, 1:50; Abcam), Snail (ab216347, 1:1000; Abcam), ZEB1 (ab203829, 1:500; Abcam), Notch1 (ab52627, 1:2000; Abcam), p21 (ab109199, 1:1000; Abcam), Hes1 (ab108937, 1:1000; Abcam), and Hey1 (ab154077, 1:3000; Abcam) at 4ºC, and subsequently incubated with respective secondary antibodies for 2 h at room temperature. After being washed with Tris-buffered saline (Sigma-Aldrich) 3 times, the bands were visualized by an enhanced chemiluminescence reagent (Beyotime, Shanghai, China), and the intensity of the blot was quantified by Image Lab 3.0 software.
Cell counting kit-8 (CCK-8) assays of cell viability
Cell proliferation was measured using the CCK-8 kit (Yeasen, Shanghai, China) as previously described34. Briefly, the cells transfected with sh-NC, sh-PKMYT1#1/2, and sh-PKMYT1#1 combined with jagged 1 were seeded into 96-well plates (2 × 104 cells/well) and cultured for 0 h, 24 h, 48 h, and 72 h, respectively, before the addition of serum-free DMEM (100 μl) containing 10 μl CCK-8 solution (Yeasen). After 2 h of incubation, the absorbance at 450 nm was read through a microplate reader (Molecular Devices, Shanghai, China).
Wound healing assays of cell migration
The migrative capability of MDA-MB-436 and MDA-MB-231 cells was detected by wound healing assays as previously described35. The transfected cells were incubated in 6-well plates (2 × 105 cells/well) for 12 h, and when the cells reached 100% confluence, cell monolayers were scraped using a 10-μl pipette tip and maintained in serum-free DMEM culture medium. The plates were washed with fresh medium to remove non-adherent cells, and images were taken at 0 h and 24 h using an inverted microscope (Olympus, Tokyo, Japan). The wound closure rate was calculated as follows: Wound closure rate = (0 h scratch width - 24 h scratch width)/0 h scratch width × 100%.
Transwell assays of cell invasion
Transwell assays were conducted to evaluate cell invasion as previous described36. The TNBC cells (1 × 105 cells/well) were incubated in 24-well Transwell chambers containing an 8 μm size porous membrane (Corning Incorporated, Corning, NY, USA). The upper chambers were coated with Matrigel (Corning Incorporated), and the lower chambers were filled with culture medium containing 10% FBS. After 24 h of incubation, the non-invading cells were carefully wiped using a cotton swab, and the invaded cells into the lower surface were fixed with paraformaldehyde and stained with 0.1% crystal violet. The invaded cells were imaged and counted through an inverted microscope.
Mouse xenograft model
All animal experiments and procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NIH) and approved by the Animal Care Ethics Committee of Wuhan NO.1 Hospital. Extensive efforts were made to ensure minimal suffering of the included animals. Ten female BALB/C nude mice (6-8 weeks of age) were purchased from Vital River Laboratory Animal Technology Company (Beijing, China). The mice were anesthetized and maintained on 3.5% and 2.0% isoflurane, respectively, and divided into two groups, the sh-NC and the sh-PKMYT1 groups (n = 5). The stably transfected 4×106 MDA-MB-231 cells by sh-NC or sh-PKMYT1#1 were dispersed by 2 mL saline and injected subcutaneously into the right flank of mice. Tumor volume was monitored every 3 days until day 24 with a caliper and calculated based on the formula: V (tumor) = 0.5 × length × width2. After 24 days, the mice were sacrificed by cervical dislocation under anesthesia with 2% isoflurane inhalation, and xenografted tumors were excised from sacrificed mice for weight measurement and then analyzed by immunohistochemistry staining.
Immunohistochemistry staining for E-cadherin and Ki67
Immunohistochemistry staining was performed to evaluate the expression of E-cadherin and Ki67 in tumor tissues. Tumor tissues were obtained from mouse xenograft mode. Paraffin-embedded tissue was cut into 5 μm thick slices that were fixed onto glass slides. Tissue sections were preincubated with 10% normal goat serum, followed by incubation with primary antibodies against E-cadherin (ab231303, 1 μg/mL; Abcam) and Ki67 (ab15580, 1 μg/mL; Abcam) overnight at 4°C. After washing with PBS, slides were incubated with secondary antibodies at 37°C for 10 min, and then cleaned with cold PBS and treated with peroxidase conjugated-biotin streptavidin complex for 10 min. Finally, stains were examined with 3,3'-diaminobenzidine and hematoxylin. Images were obtained using a light microscope (Olympus).
Hematoxylin and eosin staining
To investigate the effect of PKMYT1 knockdown on metastasis, the lungs were obtained from xenograft models and fixed in 10% formalin buffer and embedded in paraffin. The tissues were then sectioned at 2 μm and stained with hematoxylin and eosin (Sigma-Aldrich). The images were taken using a light microscope (Olympus).
Statistical analysis
Independent experiments were performed at least 3 times. The data were analyzed using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) and expressed as the mean ± standard deviation. All analyses were performed depending on the distribution (parametric or non-parametric) of the data. The normality distribution and the homogeneity of variance were evaluated using the Shapiro–Wilk test and the Levene's test, respectively. p > 0.05 indicated that the assumption of normality of data and homogeneity of variance was consistent, and further, parameter testing could be performed. One-way analysis of variance (ANOVA) followed by the Tukey's post hoc analysis was used to compare differences among three or more groups, whereas two-group comparisons were performed using the Student's t-test. p < 0.05 was considered statistically significant.
RESULTS
PKMYT1 is upregulated in invasive breast cancer tissues and metastatic cell lines
To investigate the role of PKMYT1 in BC progression, the expression of PKMYT1 in invasive breast cancer was predicted by the TCGA database on the UALCAN website. As shown in Fig. 1A, PKMYT1 expression is upregulated in luminal, HER2-positive, and triple-negative BC tissues compared with normal breast tissue samples. As Kaplan–Meier plotter analysis revealed, patients with high PKMYT1 expression exhibited a remarkably shorter recurrence-free survival than those with low PKMYT1 expression (Fig. 1B). Then, RT-qPCR analysis was performed on 50 pairs of surgical specimens (tumor and adjacent non-tumor tissue samples) collected from patients newly diagnosed with BC. We found a significant increase in PKMYT1 expression in luminal, HER2-positive, and triple-negative BC tissue samples as compared to paired non-tumor tissue samples (Fig. 1C). The correlation between PKMYT1 expression and various clinicopathological parameters was analyzed, and the results are shown in Table 1. We found that high PKMYT1 expression was significantly associated with lymph node metastasis, histological grades, pTNM stages, and recurrence/metastasis, but there was no significant difference among the age, tumor size, and molecular subtypes. Finally, the expression of PKMYT1 in MCF-10A, MCF7, T47D, MDA-MB-436, and MDA-MB-231 was detected by RT-qPCR analysis and Western blotting. The results showed that the mRNA and protein levels of PKMYT1 were remarkably upregulated in BC cells, and PKMYT1 expression was higher in MDA-MB-436 and MDA-MB-231 cells than other two BC cell lines (Fig. 1D-E). These results demonstrate that PKMYT1 is upregulated in BC tumors and cells.

***p < 0.001.
BC: Breast cancer; SD: standard deviation.
Figure 1. PKMYT1 is upregulated in BC tissues and cells. A: the expression of PKMYT1 in breast invasive cancer tissue samples was predicted by the UALCAN website (http://ualcan.path.uab.edu/). B: Kaplan–Meier plotter analysis (http://kmplot.com/analysis/index.php?p=service&cancer=breast) of the recurrence-free survival of patients with high and low PKMYT1 expression. C: the expression of PKMYT1 in luminal, HER2-positive, and triple-negative BC tissues and paired normal adjacent samples was measured by RT-qPCR. D-E: the mRNA and protein levels of PKMYT1 in BC cell lines (MCF7, T47D, MDA-MB-436, and MDA-MB-231) and normal breast epithelial cell line (MCF-10A) were measured by RT-qPCR and Western blotting. Data are expressed as the mean ± SD of three independent experiments.
PKMYT1 knockdown inhibits cell migration, invasion, and epithelial-mesenchymal transition process in triple-negative breast cancer
To explore the functions of PKMYT1 in TNBC, sh-PKMYT1#1/2 was used to knock down PKMYT1 expression in MDA-MB-436 and MDA-MB-231 cells, and their knockdown efficiency was confirmed by the experimental results of RT-qPCR and Western blotting (Fig. 2A-B). Then as shown by CCK-8 assays, the viability of MDA-MB-436 and MDA-MB-231 cells was significantly reduced by PKMYT1 depletion (Fig. 2C). Next, the results of wounding healing assays and Transwell assays revealed that PKMYT1 knockdown limited the migrative and invasive capabilities of MDA-MB-436 and MDA-MB-231 cells (Fig. 2D-E). These results suggest that PKMYT1 knockdown can inhibit the proliferation, migration, and invasion of TNBC cells.
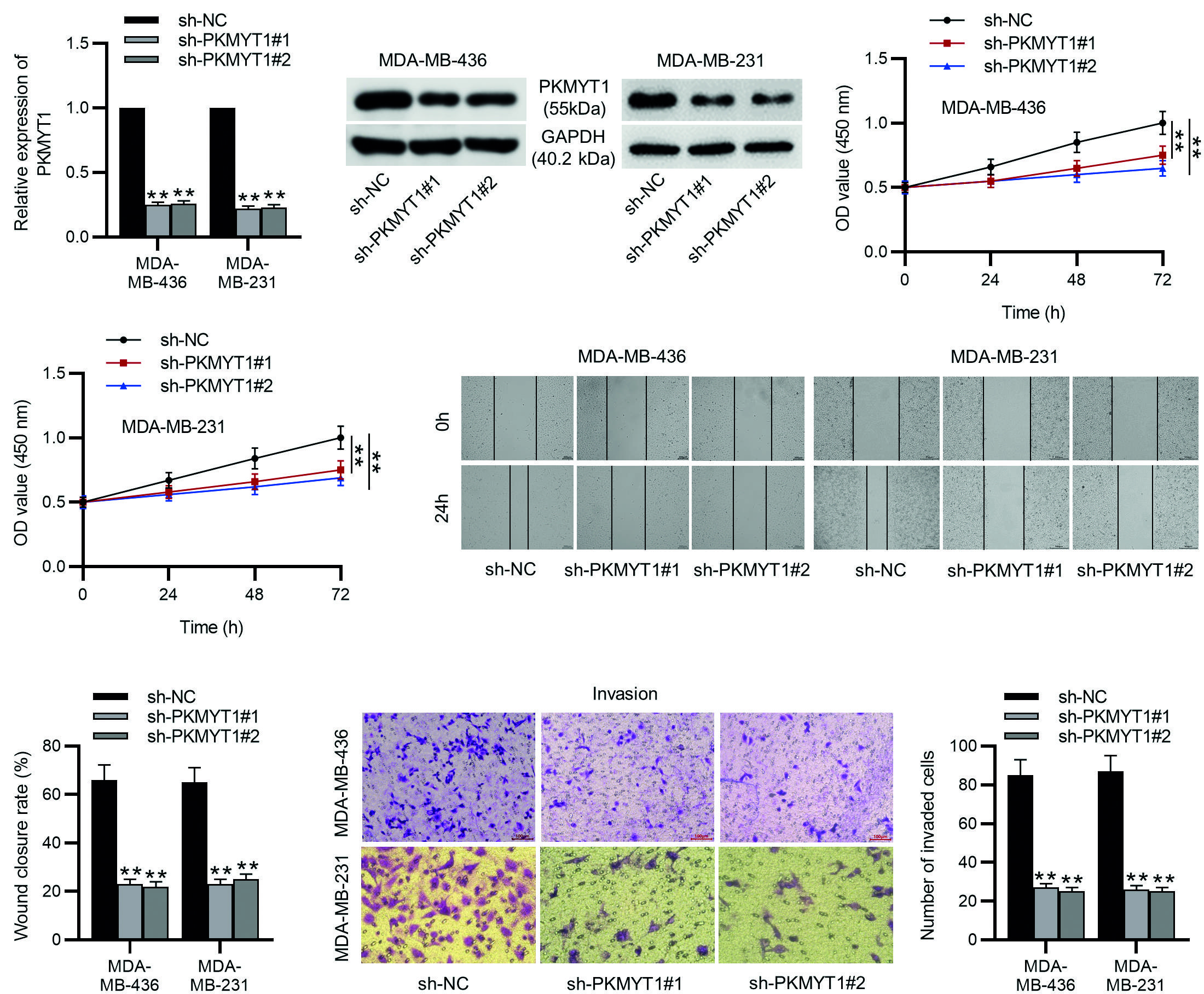
**p < 0.01.
TNBC: triple-negative breast cancer; SD, standard deviation; sh-PKMYT1, short hairpin RNA targeting PKMYT1; NC, negative control.
Figure 2. PKMYT1 knockdown inhibits TNBC cell proliferation, migration, and invasion. A, B: the MDA-MB-436 and MDA-MB-231 cells were transfected with sh-NC, sh-PKMYT1#1, or sh-PKMYT1#2 for 48 h. RT-qPCR and Western blotting were conducted to determine the knockdown efficiency of sh-PKMYT1#1/2. C: the viability of MDA-MB-436 and MDA-MB-231 cells after indicated transfection was assessed by CCK-8 assays. D: cell migration in different groups was examined by wound healing assays. E: cell invasion was assessed by Transwell assays. Data are expressed as the mean ± SD of three independent experiments.
PKMYT1 silencing inhibits EMT
EMT is a critical event for cancer metastasis, and migration and invasion are important features of the EMT37. Consequently, we detected the protein levels of EMT markers in TNBC cells. As Western blotting demonstrated, the MDA-MB-436 or MDA-MB-231 cells transfected with sh-PKMYT1#1/2 exhibited higher E-cadherin protein levels and lower N-cadherin, Snail, and ZEB1 protein levels than the sh-NC-transfected TNBC cells (Fig. 3A-C). These results support that PKMYT1 knockdown prevents the metastatic potentials of TNBC cells through the inhibition of EMT event.
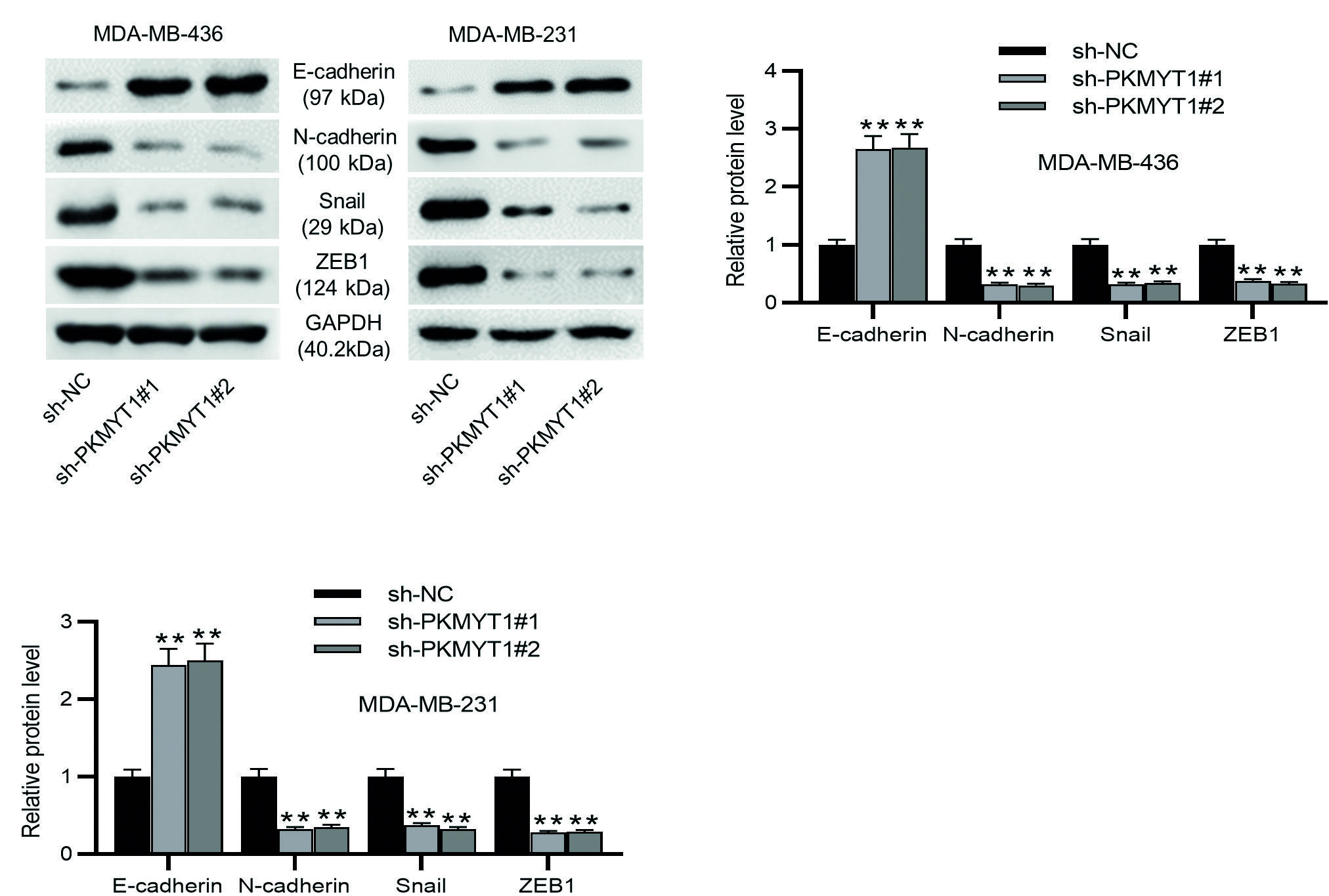
**p < 0.01.
TNBC: triple-negative breast cancer; SD: standard deviation; sh-PKMYT1: short hairpin RNA targeting PKMYT1; NC: negative control; EMT: epithelial-mesenchymal transition.
Figure 3. PKMYT1 knockdown inhibits EMT in TNBC cells. A-C: the levels of EMT-related proteins (E-cadherin, N-cadherin, Snail, and ZEB1) were evaluated by Western blotting. Data are expressed as the mean ± SD of three independent experiments.
Notch signaling is inactivated by PKMYT1 depletion
To investigate the function of PKMYT1 in the Notch pathway, we measured the protein levels of Notch1, p21, Hes1, and Hey1 by Western blotting. The results demonstrated that PKMYT1 knockdown led to significant decreases in the protein levels of Notch1, p21, Hes1, and Hey1 in MDA-MB-436 and MDA-MB-231 cells (Fig. 4A-C). All these statistics demonstrate that PKMYT1 knockdown inhibited the activation of the Notch pathway.

**p < 0.01.
TNBC: Triple-negative breast cancer; SD: standard deviation; sh-PKMYT1: short hairpin RNA targeting PKMYT1; NC: negative control.
Figure 4. Notch signaling is inactivated by PKMYT1 depletion in TNBC cells. A-C: Western blotting was performed to measure the levels of Notch-associated proteins, including Notch1, p21, Hes1, and Hey1 in MDA-MB-436 and MDA-MB-231 cells transfected with sh-NC or sh-PKMYT1#1/2. Data are expressed as the mean ± SD of three independent experiments.
Notch activation abolishes the sh-PKMYT1-mediated inhibition in triple-negative breast cancer cell proliferation, migration, and invasion
Whether the effect of PKMYT1 in the development of TNBC was mediated by Notch signaling which was determined by rescue experiments. Jagged 1 (a Notch activator) was added into TNBC cells after intervening PKMYT1 expression, and the pathway activating role of jagged 1 in Notch signaling was verified by RT-qPCR and Western blotting (Fig. 5A-B). Then, CCK-8 assays, wound healing assays, and Transwell assays were performed to detect the role of jagged 1 in the proliferation, migration, and invasion in MDA-MB-436 and MDA-MB-231 cells transfected with sh-PKMYT1#1. The results showed that jagged 1 reversed the inhibitory effect of PKMYT1 knockdown on cell proliferation, migration, and invasion (Fig. 5C-E). These results demonstrate that activation of Notch signaling abolished the inhibitory effect of PKMYT1 knockdown on cell proliferation, migration, and invasion in TNBC.

*p < 0.05.
**p < 0.01.
***p < 0.001.
TNBC: triple-negative breast cancer; SD: standard deviation; sh-PKMYT1: short hairpin RNA targeting PKMYT1.
Figure 5. Notch activation abolished the inhibitory effects of PKMYT1 silencing on the proliferation, migration, and invasion of TNBC cells. A, B: MDA-MB-436 and MDA-MB-231 cells stably expressed sh-PKMYT1#1 were treated with a specific activator of Notch signaling, jagged 1. The effects of jagged 1 on Notch signaling were detected by RT-qPCR and Western blotting. C: CCK-8 of cell viability. D: wound healing assays of cell migration. E: Transwell assays of cell invasion. Data are expressed as the mean ± SD of three independent experiments.
Notch activation limits the repressive effect of PKMYT1 knockdown on epithelial-mesenchymal transition
As shown by Western blotting, jagged 1 treatment abolished the sh-PKMYT1#1-mediated increase in the protein levels of E-cadherin and decrease in the protein levels of N-cadherin, Snail and ZEB1, indicating that activation of Notch signaling impaired the suppressive effect of PKMYT1 knockdown on EMT (Fig. 6A-C). These findings suggest that PKMYT1 can promote EMT by activating Notch signaling.

**p < 0.01.
***p < 0.001.
TNBC: Triple-negative breast cancer; SD: standard deviation; sh-PKMYT1: short hairpin RNA targeting PKMYT1; EMT: epithelial-mesenchymal transition.
Figure 6. Notch activation limits the suppressive effect of PKMYT1 knockdown on EMT in TNBC cells. A-C: Western blotting was performed to measure the levels of EMT-related proteins (E-cadherin, N-cadherin, Snail, and ZEB1). Data are expressed as the mean ± SD of three independent experiments.
PKMYT1 knockdown inhibits tumorigenicity and metastasis in vivo
To further assess the anti-tumor effect of PKMYT1 downregulation on tumor growth and metastasis, we established a xenograft tumor model by subcutaneously injecting PKMYT1-silencing MDA-MB-231 cells or sh-NC-transfected MDA-MB-231 cells into the right flank of mice. Representative images of tumor xenografts isolated from nude mice on Day 24 are shown in Fig. 7A. Compared with the scramble shRNA control group, the volume and weight of PKMYT1 shRNA group were significantly decreased (Fig. 7B-C). Then, immunohistochemistry staining of E-cadherin and Ki67 (a cell proliferation marker) was performed. The results showed that PKMYT1 knockdown increased E-cadherin expression while decreasing Ki67 expression (Fig. 5D). Moreover, as shown by hematoxylin and eosin staining, metastasis to the lungs was inhibited by PKMYT1 knockdown (Fig. 7E). Collectively, these results indicate that PKMYT1 knockdown inhibits tumorigenesis and metastasis of TNBC cells in vivo. Figure 8 presents the schematic diagram depicting the mechanisms by which PKMYT1 regulates cancer cell proliferation, migration, and invasion and EMT. PKMYT1 activates the Notch pathway to promote EMT, proliferation, migration, and invasion of TNBC cells.

**p < 0.01.
SD: Standard deviation; sh-PKMYT1: short hairpin RNA targeting PKMYT1; NC: negative control.
Figure 7. PKMYT1 knockdown inhibits tumorigenicity and metastasis in vivo. A: MDA-MB-231 cells transfected with sh-NC or sh-PKMYT1#1 were subcutaneously injected into BALB/C nude mice. Tumors were excised from nude mice at day 24. Representative images of tumor xenografts were presented. B: tumor growth curves. C: tumor weight. D: immunohistochemistry staining of E-cadherin and Ki67. E: representative images for hematoxylin and eosin staining of lung tissues. Data are expressed as the mean ± SD. N = 5.
DISCUSSION
At present, surgery supplemented by chemotherapy results in an increase in survival rate. However, delayed diagnosis, drug resistance, and recurrence possibility of TNBC load pressure on clinical treatment. Aberrations in gene expression are linked to TNBC pathogenesis38. In the present study, we investigated the biological functions of PKMYT1 and its related mechanisms in the development of TNBC.
Metastasis is highly complex and involves multiple cellular mechanisms including cell invasion and migration39. PKMYT1 is suggested to facilitate cancer progression by promoting cancer cell migration and invasion. For example, transcriptional activation of PKMYT1 augments cell motion in lung cancer40. In addition, PKMYT1 functions as a tumor promotor in osteosarcoma as evidenced by increased cell invasion and migration following PKMYT1 overexpression41. Moreover, PKMYT1 functions as an oncogene in colorectal cancer that enhances cell migration and invasion42. Furthermore, PKMYT1 downregulation inhibits the colony-formation, proliferation, migration, and invasion and promotes the apoptosis of TNBC cells25. In the present study, we found that PKMYT1 depletion facilitated TNBC cell migration and invasion, similar to the previous findings. In this study, in vivo results revealed that PKMYT1 knockdown inhibited tumor growth and lung metastasis.
During cancer development, epithelial cancer cells are conferred with migrative and invasive features by EMT. The epithelial-related molecule E-cadherin, mesenchymal marker N-cadherin, and several EMT regulating transcription factors, including Snail and ZEB1, are involved in the EMT process43. Recent studies illustrate that PKMYT1 knockdown can prevent the EMT phenotype in renal cell carcinoma44, oral squamous cell carcinoma45, esophageal squamous cell carcinoma46, and hepatocellular carcinoma47. In the present study, we found that PKMYT1 knockdown increased E-cadherin protein levels and decreased N-cadherin, Snail, and ZEB1 protein levels in MDA-MB-436 and MDA-MB-231 cells, illustrating that PKMYT1 depletion inhibited EMT of TNBC cells.
Notch signaling promotes metastatic phenotypes in TNBC cells48,49. Upregulation of PKMYT1 can enhance lung cancer progression by activating the Notch signaling32. This study revealed that PKMYT1 downregulation decreased the protein levels of Notch1, p21, Hes1, and Hey1, indicating that PKMYT1 knockdown inactivated Notch signaling. Jagged 1 is recognized as a specific activator of Notch signaling. The present study demonstrated that Jagged 1 treatment reversed the suppressive effects of PKMYT1 knockdown on the malignant phenotypes of TNBC cells.
There are limitations to this study. First, more molecular mechanisms by which PKMYT1 promote TNBC development are needed to be explored. Second, additional methodologic and clinical validations are warranted to elucidate the role of PKMYT1 as a target for TNBC diagnosis. Finally, it is possible that analysis of a larger cohort of patients with TNBC could influence several results which approached but failed to reach statistical significance. Further studies are required to address the abovementioned limitations.
In conclusion, this study demonstrates that PKMYT1 can aggravate cell migration and invasion and EMT phenotype in TNBC through activation of the Notch signaling. These results indicate that PKMYT1 is essential in the regulation of TNBC progression, suggesting an experimental basis for its use as a cancer biomarker and therapeutic target for TNBC treatment, and expands its potential clinical value.











 nueva página del texto (beta)
nueva página del texto (beta)