INTRODUCTION
Suicide is one of the leading causes of death worldwide1 and a growing health problem, as suicide rates have been increasing in many countries, including Mexico2. Suicide is a complex behavior where several contributors interact, i.e., genetic, environmental, and sociocultural factors3,4. Among suicide-related factors, epigenetic changes seem to be particularly relevant in suicidal behavior, due to their mediating role in the complex interaction between genotype and environment5,6. Epigenetic modifications, such as DNA methylation, may regulate gene expression patterns in the brain, leading to the neurobiological abnormalities associated with suicidal behavior7.
Alterations in DNA methylation and gene expression have been widely evidenced in the suicidal brain and have been implicated in suicide pathology8-16. However, our understanding of the regulatory effect of DNA methylation in gene expression in suicides is scarce, since correlations between DNA hypermethylation and gene expression have been limited to candidate genes, such as tropomyosin-related kinase B (TRKB), glucocorticoid receptor (GR), ornithine decarboxylase antizymes 1 and 2 (OAZ1 and OAZ2), quaking homolog KH domain RNA binding (QKI), S-adenosylmethionine decarboxylase (AMD1), and arginase 2 (ARG2)17-21.
The integration of molecular traits at different levels is crucial to the elucidation of mechanisms leading to observed gene expression alterations in the suicidal brain. In this study, we aimed to identify CpG sites whose methylation levels are correlated with gene expression at a genome-wide level in the prefrontal cortex of suicides. The prefrontal cortex was selected for its analysis due to its importance in suicide, as there is extensive evidence of structural, cognitive, and molecular alterations in this brain area in individuals with suicidal behavior22-26.
The relevance of the bi-directional interactions between DNA methylation and transcription factors (TF) for regulating gene expression is noteworthy, as DNA methylation may inhibit TF binding and the interaction of TF with methylated CpG sites may translate DNA methylation signals into biological actions27,28. In this study, we performed a prediction of TF binding sites (TFBS) located in promoters related to the identified CpG sites. These are the preliminary results of an ongoing study on suicide genomics at the National Institute of Genomic Medicine in Mexico.
METHODS
Samples
Brain tissue samples from 21 male suicides and six male non-suicides were analyzed in this study. Male suicide samples have previously been described in Rodríguez-López et al.29, where the influence of genetic variants in gene expression and methylation profile in suicides was studied. Postmortem brain samples were collected at the Institute of Forensic Sciences in Mexico City, Mexico. Suicides died by self-inflicted injuries, and non-suicides presented a sudden non-suicidal self-inflicted death, as determined by the coroner.
Postmortem interval, which is the interval between the estimated time of death and sample collection, ranged from 8.31 to 23 h in the analyzed sample. Age and postmortem interval differences between suicides and non-suicides were evaluated by Wilcoxon rank-sum test. All individuals included in this study were Mexico City residents.
The present study was approved by the Ethical Committee for Human Research at the National Institute of Genomic Medicine and the Bioethics Committee of the Faculty of Medicine at the National Autonomous University of Mexico (UNAM). Tissue samples from the dorsolateral prefrontal cortex (Brodmann area 9) were sectioned during an autopsy protocol, following a previously described procedure30. Samples were stored in RNAlater at -80°C until their use.
A consensus diagnosis of psychiatric and neurological disorders was made for each individual with information obtained from the coroners records. Information contained in the coroners records included (a) demographic information; (b) the autopsy report; (c) description of death circumstances; (d) results from the toxicology test, performed in both peripheral blood and brain tissue as part of the autopsy protocol; (e) police reports; (f) medical and psychiatric notes, if available; (g) suicide notes; (h) testimonies from relatives and witnesses of suicide completers; and (i) the death certificate. Consensus diagnosis was performed according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) criteria31.
Nucleic acids isolation
Genomic DNA was isolated from ~100 mg of tissue with the QIAamp DNA Mini Kit from Qiagen. Meanwhile, RNA isolation was performed using the RNeasy kit from Qiagen. Isolation of both nucleic acids was performed according to the manufacturers instructions. Then, DNA and RNA quantity and integrity were determined using the NanoDrop 2000 spectrometer (Thermo Fisher, Wilmington, DE, USA) and agarose gel electrophoresis, respectively. In addition, the RNA integrity number (RIN) of the samples was evaluated using Agilent Bioanalyzer. All samples were further processed as they complied with standard quality parameters, such as an adequate concentration (> 50 ng/µl) and integrity. RNA samples had an RIN > 6.
Microarray detection
Isolated DNA was bisulfite-converted using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) following the manufacturers protocol. Then, bisulfite-converted DNA was hybridized into the Illumina Infinium Human Methylation 450K BeadChip (Illumina, San Diego, CA, USA) to evaluate the methylation profile of the samples.
For gene expression profiling, RNA samples were labeled into biotinylated cRNAs with the TargetAmp-Nano Labeling Kit (Epicentre). Then, samples were hybridized into the Illumina HumanHT-12 v4 BeadChip (Illumina, San Diego, CA, USA) microarray. The processing of both arrays was performed according to the established manufacturers conditions and protocol and was scanned on an iScan Microarray Scanner (Illumina, San Diego, CA, USA) immediately after their respective protocol.
DNA methylation data pre-processing
IDAT files containing raw DNA methylation data were processed using minfi quality control and normalization options32. Microarray probes were filtered according to the following criteria: (i) detection p > 0.05; (ii) with at least one SNP of minor allele frequency (MAF) > 5%, according to the allele frequency estimated from 1000 Genomes Project samples33; and (iii) probes previously reported as cross-reactive33. A matrix of beta values was generated for correlation analysis.
Gene expression data pre-processing
Data pre-processing was conducted in the Gene Expression Module of GenomeStudio v2.0 software (Illumina, USA). Microarray probes with a detection p > 0.05 in more than 5% of the arrays were filtered out. Then, raw probe intensities were normalized and log(2)-transformed with the quantile method34. A matrix of log(2) values was generated for correlation analysis. After quality control and pre-processing steps in each platform, we obtained a total of 8670 expressed genes and 468,922 CpG methylation sites, which were further analyzed for correlation between both molecular traits.
Correlation analysis
Correlation between DNA methylation beta values and gene expression values was tested with a linear regression adjusted by age and postmortem interval implemented in the R package MatrixEQTL35. To evaluate the effect of DNA methylation in both enhancers and promoters, distance thresholds for detection of CpG-gene pairs were set at 1 Mb, 100 kb, and 50 kb upstream or downstream from the transcription start site of the corresponding genes. A significant correlation was defined when its two-sided p- < 0.05 after correction for multiple testing with the Benjamini-Hochberg False Discovery Rate (FDR) method36. Then, the correlation of the CpG sites correlated with gene expression in the suicide group was tested in the non-suicide control group.
To evaluate whether the gene expression-methylation correlations found in the suicide group were related to a comorbid psychiatric disorder, an additional correlation analysis was performed, separating suicides with major depression disorder (n = 8), which was the most common mental disorder in our sample, and suicides without mental comorbidities (n = 10).
CpG sites were annotated with the IlluminaHumanMethylation450kmanifest package37. To gain insight into the key processes where genes correlated with CpG methylation are participating, gene ontology (GO) enrichment analysis was performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID), version 6.838,39. GO terms with a modified Fisher exact p < 0.05 (EASE score) were considered enriched40.
Prediction of TFBS
First, the DNA sequences from the CpG sites correlated with gene expression were obtained from the UCSC Genome Browser available at http://genome.ucsc.edu/41. CpG site sequences were used as input in the Neural Network Promoter Prediction tool (NNPP) v2.2 (http://www.fruitfly.org/ index.html) to find possible transcription promoters and their sequence42. Second, a TFBS prediction was performed in PROMO (http://alggen.lsi.upc.es/cgi-bin/promo) v8.3 for each transcription promoter sequence43,44. The search was restricted to eukaryotic factors and sites. Third, we evaluated the presence of gene expression changes in the TF with binding activity to the predicted TFBS in pre- and post-natal developmental periods with the query of developmental transcriptome reported in the BrainSpan database (www.brainspan.org)45. To identify the specific stage where gene expression changes take place, additional comparisons between the following stages were performed: prenatal period (0-38 weeks), infancy (birth-18 months), childhood (19 months-11 years), adolescence (12-19 years), and adulthood (≥20 years) in all brain regions.
RESULTS
Data from 21 male suicides and six male non-suicide controls were analyzed in this study. No significant differences were found between suicides and non-suicides in terms of age (p = 0.5852) and postmortem interval (p = 0.5046). The sample characteristics are summarized in table 1. A total of 22 CpG sites showed a significant correlation with gene expression after correction for multiple testing in the suicide group. Among these, 17 pairs (77.27%) were located within 1 Mb upstream or downstream from the transcription start site. Meanwhile, three were found within 100 kb and two within 50 kb from the transcription start site.
Table 1 Characteristics of subjects
| Suicides | Non-suicides | |
|---|---|---|
| Number of individuals | 21 | 6 |
| Age (years) | 28.4 ± 9.15 | 29.33 ± 8.09 |
| Postmortem interval (hours) | 14.33 ± 4.73 | 15.53 ± 3.78 |
| Psychiatric disorder* | 8/3/10 | 0/1/5 |
| Substance reported in toxicology test** | 9/1/11 | 0/0/6 |
| Mechanism of death† | 18/0/2/1 | 2/4/0/0 |
Values indicate number of subjects unless otherwise indicated. Continuous data are presented as mean ± standard deviation.
*Major depression disorder/Personality disorder/None.
**Alcohol/Cocaine/None.
†Asphyxia/Gunshot/Puncture wound/Trauma.
Regarding location relative to genes, six CpG sites were located in promoter regions and three in enhancer regions. Regarding the location of CpG sites relative to CpG islands, 12 sites (54.54%) were located inside CpG islands, six sites (27.27%) on island shelves (four in north and two in south shelf), and four sites (18.19%) were located on island shores (three in north and one in north shore). The list of CpG-gene pairs is shown in table 2.
Table 2 Significantly correlated CpG-gene pairs
| Chromosome | Gene symbol | Gene coordinates* | CpG site | CpG coordinates** | Enhancer/Promoter | Relation to CpGi† | Beta | FDR‡ |
|---|---|---|---|---|---|---|---|---|
| cis distance 1 Mb | ||||||||
| 7 | C7ORF38 | 99143923-99156115 | cg05313743 | 99613104-99613797 | Promoter | Island | 10.3083 | 0.01635 |
| 10 | NKX6-2 | 134598320-134599537 | cg06715410 | 134411241-134411481 | N-Shelf | 8.55859 | 0.04008 | |
| cg00862314 | 134359533-134359780 | Promoter | N-Shore | 7.8911 | 0.04008 | |||
| PAOX | 135192741-135205198 | cg14657458 | 134468815-134469082 | Island | 8.27878 | 0.04008 | ||
| cg00470761 | 134597357-134602649 | N-Shore | 7.85877 | 0.04008 | ||||
| cg06715410 | 134411241-134411481 | N-Shelf | 8.45828 | 0.04008 | ||||
| C10ORF92 | 134621896-134756089 | cg11518184 | 133849676-133850826 | Enhancer | Island | 7.807059 | 0.04008 | |
| DNAJB12 | 74092588-74114907 | cg06185830 | 74113815-74114792 | Promoter | Island | 7.850115 | 0.04008 | |
| PIP5K2A | 22823766-23003503 | cg17272579 | 22604907-22605632 | S-Shelf | 7.894761 | 0.04008 | ||
| 11 | ATPGD1 | 67183149-67193078 | cg20158826 | 66360097-66360834 | N-Shore | 10.0909 | 0.03481 | |
| 15 | BBS4 | 72978520-73030817 | cg18517195 | 71184276-71184689 | Island | 8.69968 | 0.02961 | |
| LOC653073 | 30375038-30386084 | cg05678749 | 27215951-27216856 | Island | 9.701103 | 0.01579 | ||
| 17 | WDR79 | 6125879-6127775 | cg13759852 | 8054550-8055835 | Enhancer | S-Shore | 10.42292 | 0.02465 |
| 18 | MBP | 74690789-74844774 | cg03731646 | 74499316-74499641 | Island | 8.995957 | 0.00751 | |
| 19 | APOC1 | 45417577-45422606 | cg27009008 | 46387327-46389578 | Promoter | Island | 14.69784 | 0.00040 |
| 21 | CBR3 | 37506614-37518860 | cg00994804 | 36258952-36259472 | Enhancer | Island | 8.643333 | 0.01656 |
| 22 | SDF2L1 | 21996542-21998588 | cg19948255 | 19572306-19572392 | Enhancer | Island | 9.202877 | 0.02523 |
| cis distance 100 kb | ||||||||
| 9 | FNBP1 | 132649458-132805473 | cg13728604 | 132630324-132630823 | N-Shelf | 7.6801 | 0.03040 | |
| 19 | AXL | 41725104-41767672 | cg08554936 | 46518283-46520080 | Promoter | Island | 7.97518 | 0.04841 |
| ZNF480 | 52800426-52829180 | cg18384778 | 57617601-57618448 | Island | 9.308521 | 0.01536 | ||
| cis distance 50 kb | ||||||||
| 11 | CTNND1 | 57520756-57586652 | cg10964211 | 57281671-57283305 | Promoter | N-Shelf | 7.73769 | 0.02597 |
| RASSF7 | 560309-564025 | cg11972598 | 597465-597700 | S-Shelf | 7.841229 | 0.02597 | ||
*Genomic location of the gene (NCBI37/hg19).
**Genomic location of the CpG site (NCBI37/hg19).
†Location relative to CpG islands.
‡FDR-adjusted p-value.
Functional analysis with DAVID indicated that five genes were significantly associated with central nervous system development (GO: 0007417, p = 0.0020). Of these genes, CTNND1 and AXL are involved in the VEGFA-VEGFR2 pathway (HSA-4420097, p = 0.0058).
A total of 416 TF was predicted for the CpG sites identified in the present study. Of these, only 9e (USF1, TBP, SF1, NRF1, RFX1, SP3, PKNOX1, MAZ, and POU3F2) showed differential expression, i.e., exhibited significantly different expression levels when comparing gene expression profile in pre- and post-natal developmental periods in the BrainSpan database. The gene expression pattern of these TF was analyzed across developmental stages, as shown infigure 1. It was noteworthy that TF expression predicted to bind to promoter regions located in CpG sites was higher during prenatal stages and decreased as development progressed. Complete results of TF gene expression analysis in BrainSpan are shown in Supplementary table 1.
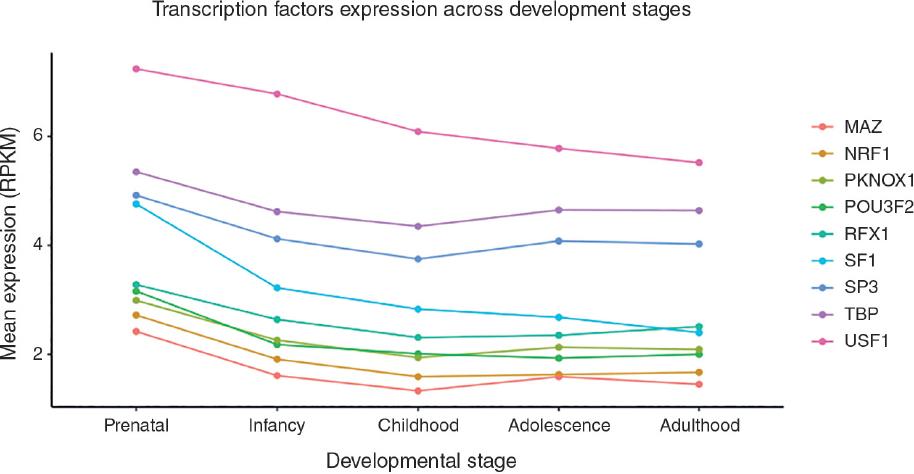
Figure 1 Mean expression levels of transcription factors predicted to bind to promoter regions located in CpG sites correlated with gene expression in the prefrontal cortex of suicides during developmental stages.
The correlation between the CpG sites correlated with gene expression in the suicide group was tested in the non-suicide control group. However, this analysis did not show any significant correlation between the tested CpG site-gene pairs. The results of the correlation test in the non-suicide group are shown in Supplementary table 2.
In addition, correlations were tested, separating suicides with major depression disorder (n=8) from suicides without mental comorbidities (n = 10). Correlations were not significant in either group. The complete results of this sub-analysis are shown in Supplementary tables 3 and 4.
DISCUSSION
To the best of our knowledge, this is the first whole-genome transcriptomic-methylomic correlation analysis in the dorsolateral prefrontal cortex of individuals who died by suicide. The previous studies have reported genes regulated by DNA methylation, mainly by candidate genes studies17-21. Correlations found in the suicide group were not detected in the non-suicide group. Thus, our results provide new insights into the effects of DNA methylation on the brain gene expression of suicide.
Among the genes whose expression was found to be correlated to CpG sites, we identified genes involved in neurodevelopment, such as BBS4, NKX6-2, AXL, CTNND1, and MBP. This finding is consistent with the previous reports of misexpression of genes involved in central nervous system development in suicide14,46. In addition, we identified a significant correlation with DNA methylation for the expression of genes involved in processes contributing to remodelings in the central nervous system, such as RASSF7 involved in cell proliferation and cytoskeletal organization47,48; CTNND1 and MBP, which have been implicated in cell adhesion and migration49; and MBP, which encodes for a major constituent of the myelin sheath of oligodendrocytes50. Furthermore, the NKX6-2 gene has shown to have an important role in the generation of cortical interneurons and their differentiation51.
In addition, the prediction of TFBS in the identified CpG sites related to TF involved in neurodevelopment suggests a role of DNA methylation-TF interactions in the gene expression patterns observed in suicides. DNA-methylation-TF interactions have been related to the downregulation of gene expression52. However, recent studies have proven that such interactions are complex, and their effects are varied. Moreover, the effects seem to be bidirectional. For example, among the predicted TF found in our results, RFX1 modulates epigenetic modifications by recruiting the histone methyltransferase SUV39H153. Meanwhile, methylation in the core-promoter region of the chondromodulin-I gene regulates the binding of transcriptional activator Sp354. Thus, the effects of DNA methylation-TF interactions in neurodevelopment should be further studied.
Furthermore, the most remarkable TF expression changes were found between prenatal and postnatal developmental stages. The vulnerability of the brain to insults during the earliest developmental stages has been acknowledged as one of the factors associated with the susceptibility to neurodevelopmental and psychiatric disorders55. Therefore, our findings suggest a possible role for TF on neurodevelopmental alterations reported in the suicidal brain.
Our results suggest a role for DNA methylation and possibly TFs as well, in the regulation of neurodevelopmental gene expression, providing insight into the molecular mechanisms that might lead to neuroanatomical abnormalities in both gray and white matter reported in individuals with suicidal behavior56-58.
Our analysis suggests that polyamine oxidase (PAOX) expression may be functionally regulated by methylation. PAOX encodes for the PAOX, an enzyme that catalyzes the oxidation of N1-acetylspermidine and N1-acetylspermine products of spermidine/spermine N1-acetyltransferase (SAT1), producing spermidine and putrescine. Spermidine and putrescine levels found in the brain of suicide completers with a history of major depression were significantly higher compared to controls59. It has been hypothesized that this polyamine accumulation may be a consequence of decreased SAT1 expression60. However, the upregulation of PAOX provides a better explanation for the reported levels of polyamines in the suicidal brain, as both of its products appear to be increased in the suicidal brain.
Another possibility is that increased PAOX expression is a compensatory mechanism that decreases these levels60. PAOX appears to be controlled by substrate availability26; thus, its regulation at a transcriptional level by methylation might be an additional regulatory mechanism, as polyamine homeostasis is a highly regulated mechanism60. Alterations in the expression of polyamine-related genes have been consistently reported in the brain of suicide completers61. The regulation by DNA methylation in polyamine biosynthetic genes, such as AMD1, ARG2, OAZ1, and OAZ2, has been previously reported21. Therefore, the role of PAOX in suicide is worthy of exploring in future research.
Recently, an abnormal telomere length was identified in suicide completers62. The protein encoded by WDR79 is crucial in telomere synthesis, as it acts as a telomerase holoenzyme component, facilitating its assembly and translocation63. The modulation of the expression of WDR79 by DNA methylation as indicated by our results suggests a candidate mechanism, underpinning telomere alterations in suicide completers. Regarding other CpG-gene pairs identified in this study, we found genes previously linked to mental disorders, such as PIP5K2A and ZNF480, that have been implicated in schizophrenia64,65. Future studies will determine whether these genes are implicated in suicide as well.
Some of the genes whose expressions were found to be correlated with DNA methylation in our study are involved in pathways previously reported as dysregulated in suicide, as in the case of neurodevelopment and polyamines. The findings of this study improve our understanding of the regulatory mechanisms that lead to previously reported abnormalities in the suicidal brain. Furthermore, our study provided novel candidates for the investigation of genes regulated by DNA methylation in completed suicides.
It is noteworthy that this study focused on the Mexican population. Most postmortem suicide studies have focused on the Caucasian population, and studies in the Latino population regarding this subject are very scarce. Since environmental factors influencing epigenetic modifications vary among different populations, and specifically Mexican Mestizo populations have shown high genomic variability66, the study of individuals with diverse ethnic backgrounds will allow the identification of population-specific alterations in the suicidal brain. The inclusion of socio-demographic and clinical variables will allow the identification of factors associated with the observed molecular traits. In addition to DNA methylation, it is important to consider the influence of other factors in gene expression, for example, other epigenetic factors such as histone modification may influence this phenotype and should be further studied.
Limitations of the present study must be acknowledged. First, the relatively small sample size and that the analysis was restricted to male individuals. Therefore, future research with larger samples and individuals of both sexes is necessary to confirm our results and their association with suicide. Second, our sample included individuals with different psychiatric disorders, which may represent a confounding factor in our analysis. Correlations were not significant when separating suicides with major depression disorder from suicides without comorbid mental disorders, as well as in non-suicides. This lack of correlation might be due to the small sample size found in each group. The tendency of a cg06715410/NKX6-2 pair to correlate with an FDR corrected p-value of 0.07793 in the non-suicide group is noteworthy. This finding encourages correlation testing in larger samples of suicides and non-suicides with and without mental disorders for identifying correlations specific to each condition. Future studies should include groups of suicides and non-suicides with mental disorders in an adequate number for estimating the effect of the comorbid mental disorder in gene expression and methylation. As part of an ongoing project, which started in 2018, we expect to collect more samples and to overcome the aforementioned limitations.











 nueva página del texto (beta)
nueva página del texto (beta)


