INTRODUCTION
Psychiatric disorders are complex diseases caused by multiple risk factors1,2. During the past decade, the discovery of genetic factors involved in the susceptibility to these disorders has increased rapidly, due to novel techniques such as genome-wide association studies (GWAS)3-6. GWAS have identified multiple loci among common genomic variants that have been associated to one or more psychiatric disorders. Methods to determine polygenic risk scores (PRSs) have been developed to summarize and use what is known about these multiple disease-associated loci as risk prediction tools7,8. PRS can be described as the summation of disorder-associated alleles across many loci, weighted by their effect sizes estimated from GWAS in one individual to predict one persons likelihood of developing a disease with a genetic component. Therefore, a high value of PRS could be translated into an increased risk of a particular disorder for having more individual risk variants, each one known to be associated with the same disorder. The calculation of how much an individual variant increases a disease risk, or how groups of variants increase the risk, derives from former GWAS9. Recent studies of PRS as risk prediction tools for a disease have examined how different complex diseases might share polygenetic backgrounds. They have also evaluated whether PRS can be used to predict particular traits or subtypes within groups of individuals who have the same diagnosis10-12.
Many epidemiological analyses have shown that psychiatric disorders such as bipolar disorder (BD), schizophrenia (SCZ), and major depressive disorder (MDD) have high comorbidity with substance use disorders (SUDs)13,14. The comorbidity between SUD and other psychiatric disorders is so high that the term dual diagnosis was created to specify comorbidity of these disorders15-17. Dual diagnosis, when it occurs, has been associated with multiple negative physical and psychosocial outcomes such as poorer quality of life, higher rates of relapse of substance use, and increased suicide risk18-20. Individuals with a dual diagnosis show increased severity of symptoms, which place them at high risk21. Although etiologic mechanisms underlying dual diagnosis have not been clearly established, some have suggested that individuals with dual diagnosis could have a greater genetic susceptibility22,23. Recently, PRSs for psychiatric disorders have been proposed to be useful for exploring the shared genetic susceptibility between psychiatric disorders and SUD, to understand the genetic basis of dual diagnosis24-26. In addition, PRSs of psychiatric disorders have been studied to find risk predictors for dual diagnosis27-30. However, one of the main limitations to this approach is that PRSs of psychiatric disorders were derived from GWAS in European populations, which have a highly homogeneous genetic background, and they have not been conducted in populations with a high degree of genetic admixture. There are nations around the world with a high degree of admixture, including the Mexican population31, which comes from the combination of several indigenous groups and a few European populations. Therefore, some researchers are concerned with the generalization of the use of PRS in non-European populations, and how the use of PRS could be translated to populations with heterogeneous genetic background32-34. In this sense, our aim was to explore the performance of PRS calculated from previous GWAS for psychiatric disorders when applied to Mexican individuals with a high degree of admixture who have been diagnosed with SCZ or BD, many of whom had a dual diagnosis.
METHODS
Participants
Target sample
All participants were recruited at the Carracci Medical Group and evaluated using the Diagnostic Interview for Genetic Studies (DIGS)35. Diagnoses were assigned using the DSM-V criteria for BD, SCZ, and SUD. A total of 192 individuals of Mexican ancestry were included in the analysis. Inclusion criteria: The participants parents and grandparents had to be of Mexican ancestry, meaning that mother, father, and four grandparents were born in Mexico; participants had to be 18 years of age or older. The group of cases consisted of 125 unrelated outpatients; 72 (49 males and 23 females) had a lifetime diagnosis of SCZ and 53 (25 males and 28 females), a lifetime diagnosis of BD. All patients were under psychiatric treatment for at least 3 weeks after this study began. For the control group, we used the same inclusion criteria and excluded individuals with BD, SCZ, MDD, SUD, anxiety disorders, or history of suicidal behavior. The control group consisted of 67 subjects with no lifetime history of any psychiatric disorder and who had no relatives with a known history of psychiatric disorders. We defined dual diagnosis as diagnosis of SCZ or BD with one or more lifetime SUDs (tobacco, alcohol, and illegal drugs)36. The prevalence of lifetime SUD (dual diagnosis) in the group of patients with SCZ was 40.28% (n = 29) and 57.72% (n = 29) in patients with BD. An overview of sociodemographic characteristics of the sample is reported in Table 1. This protocol was approved by the ethics and investigation committee of the National Institute of Genomic Medicine under the approval number 23/2015/I. All participants were informed of the aims of the study and gave their written consent before the study began. All protocols were performed the following guidelines of the Helsinki Declaration.
Table 1 Summary of sociodemographic data
| Sociodemographic characteristic | Bipolar disorder (n = 53) | Schizophrenia (n = 72) | Control (n = 67) |
|---|---|---|---|
| Age (years, ± SD) | 37.58 (14.03) | 33.53 (9.98) | 34.37 (12.51) |
| Years of education (years, ± SD) | 12.49 (4.03) | 10.53 (3.54) | 13.16 (5.22) |
| Gender | |||
| Male (n, %) | 25 (47.17) | 49 (68.06) | 26 (38.81) |
| Female (n, %) | 28 (52.83) | 23 (31.94) | 41 (61.19) |
| SUDs | |||
| No SUD | 24 (45.28) | 43 (59.72) | 67 (100.00) |
| Dual diagnosis | 29 (57.72) | 29 (40.28) | 0 (0.00) |
| Alcohol | 23 (43.40) | 21 (29.17) | 0 (0.00) |
| Tobacco | 29 (54.72) | 29 (40.28) | 0 (0.00) |
| Cocaine | 8 (15.09) | 1 (1.39) | 0 (0.00) |
| Cannabis | 5 (9.43) | 9 (12.50) | 0 (0.00) |
| Inhalants | 3 (5.66) | 1 (1.39) | 0 (0.00) |
| Stimulants | 3 (5.66) | 1 (1.39) | 0 (0.00) |
Dual diagnosis was defined as having a psychiatric disease diagnosis (BD or SCZ) and at least one SUD. SUDs included abuse or dependence of alcohol, cocaine, cannabis, inhalants, and/or stimulants. There were no other SUDs in this sample. SD: Standard deviation, No SUD: Patients without dual diagnosis. BD: Bipolar disorder, SUD: Substance use disorder, SCZ: Schizophrenia.
Discovery samples
As discovery samples, we used the publicly available summary statistics from GWAS to obtain the single-nucleotide polymorphisms (SNPs) and associated effect sizes, minor allele frequencies, and effect alleles, to be included in the PRSs calculation for each disorder. Data from these discovery samples came from the Psychiatric Genomics Consortium (PGC)37 and could be accessed from https://www.med.unc.edu/pgc/results-and-downloads. Data in the PGC portal are summary statistics derived from GWAS previously performed in the following disorders: autism spectrum disorders (ASD)38, attention-deficit/hyperactivity disorder (ADHD)39, BD3, MDD40, and SCZ41.
Procedures
Genotyping and imputation of target sample
DNA was extracted from peripheral leukocytes using a salting-out commercial protocol, following the specifications established by the provider (Qiagen, USA). Genotyping was performed using the whole-genome genotyping platform PsychArray BeadChip (Illumina, USA) following the protocol and conditions established by the provider. PsychArray includes approximately 560,000 polymorphisms distributed across the whole genome, as well as some variants previously associated with diverse mental psychiatric disorders including BD and SCZ. As quality control (QC), we removed all the SNPs with a minor allele frequency (MAF) of 5%, a HardyWeinberg equilibrium (HWE) p < 0.00001 for a Chi-square test; additionally, polymorphisms with a genotyping rate lower than 95% were removed. The genotyped database is available as supplementary information 1.
After the genotyping process, we performed an imputation using Beagle software; the 1000 Genomes database was utilized as reference42-44. For the following analyses, we included only SNP with a Chi-square test p-value for an HWE lower than 0.00001, a MAF higher than 0.05, and an allelic R2 higher than 0.445. After imputation and QC filter, we obtained a total of 4,835,917 SNPs.
Analyses
Polygenic risk score calculation
PRSs are measures developed to reduce the calculation of risk profiles due to polygenicity in complex diseases to a simpler and more manageable, single score for everyone. The PRSs for one individual are the summation of GWAS associated alleles to a disorder/trait, weighted by their effect sizes46. To calculate the individual PRS, we used summary statistics based on published GWAS from the Psychiatric Genomic Consortium (data free to download from: https://www.med.unc.edu/pgc/results-and-downloads) utilizing the algorithm implemented in PRSice9. PRSice calculated PRS using two sets of data, one discovery and one target sample. The discovery sample is the summary statistics from a GWAS for a specific trait or disease, with enough power to detect an association at genome-wide significance (i.e., data downloaded from GWAS and meta-analysis reported by the PGC). The target sample is the sample where PRSs are going to be calculated (our target sample was the genotype data obtained after genotyping, genotyping QC, imputation, and imputation QC).
Once we had the discovery and the target samples, PRSice performed two steps: first, the clumping process, where polymorphisms in linkage disequilibrium (LD) between the associated loci in the target sample and the discovery sample were unified47. In this analysis, we used the following clumping criteria: 250 kb and pairwise LD R2 < 0.1. Second, PRSice calculated the individuals PRS using different p-values thresholds for the associated variants in the discovery sample (with a lower bound of p = 0.0001 and an upper bound of p = 0.5; increments of 0.00005, which generated 9999 different thresholds). After the different thresholds were performed, the best-fitted model estimates were reported. This high-resolution approach allowed us to calculate the best-fitted PRS for the target sample. In this study, we report the PRS for the best estimates of models. All the models were adjusted by age, gender, and the first five principal components of global ancestry; for p-value multiple testing corrections, we performed 1000 permutations tests. The global ancestry estimation was performed with principal components analysis implemented in the GCTA software48, using a panel of 200 ancestry informative markers previously reported to reach at ancestry estimations in American populations49. For ancestry calculation, the following populations were used as references: Utahs residents with northern and western European ancestry (CEU), Yoruba residents in Ibadan from a Nigeria population (YRI) reported in the 1000 Genomes Project44, and 25 individuals of Mexican Amerindian (MA) ancestry genotyped with the multiethnic genotyping array (Illumina, San Diego, CA, USA). Once we calculated the best-fitted PRS, we compared the mean of these PRSs for psychiatric disorders between cases (subjects with BD and SCZ) and controls using a Welch t-test and considered a significant association when p < 0.05.
Calculation of SUD correlation with psychiatric diseases PRSs
One common application of PRS is to test for common genetic variation shared by two different traits or disorders46. To determine whether any of the psychiatric PRS (out of those for ADHD, ASD, MDD, SCZ, and BD) calculated with the previous algorithm, had a shared genetic etiology with a dual diagnosis phenotype within our sample, we performed a Nagelkerke test implemented in PRSice50,51. PRSice reports the variance explained by PRS in the analyzed phenotype, calculated as the difference in the Nagelkerkes pseudo-R2 from a model including the score and covariates versus a model including only the covariates. In these correlation tests, we recorded the phenotype of dual diagnosis as cases (29 individuals with both SUD and BD and 29 individuals with both SUD and SCZ) and defined a non-SUD phenotype consisting of all individuals without SUD diagnoses (67 individuals with either BD or SCZ who did not have a comorbid SUD and the 67 controls). Furthermore, for these analyses, we used the best-fitted models as described above. After finding which PRS explained a higher variance with SUD, we performed a Welch t-test comparison of this PRS between patients diagnosed with one psychiatric disease only (BD or SCZ without SUD) and patients with dual diagnosis (BD or SCZ with SUD) and considered a significant association when p < 0.05.
Effect of global ancestry estimation on the best-fitted PRS
To establish whether ancestry influences the PRS that shared a genetic effect with dual diagnosis, we performed a Spearman correlation test implemented in R52. We compared the global ancestry principal component 1 (PC1) and principal component 2 (PC2) with the PRS (best-fitted PRS and covariants adjusted). We only included PC1 and PC2 because these two global ancestry principal components divide individuals into two main populations31. We considered a correlation with PRS and global ancestry component if p < 0.05.
RESULTS
In the analysis of PRS for psychiatric disorders, we found that the only PRS that showed statistical differences between psychiatric cases (72 subjects diagnosed with SCZ and 53 subjects diagnosed with BD) and controls (67 subjects) was the PRS for MDD. A summary of the means of each PRS for ADHD, ASD, BD, MDD, and SCZ is reported in Table 2.
Table 2 Comparisons of psychiatric disorders polygenic risk scores between patients diagnosed with psychiatric disorders and controls
| Polygenic risk score | Cases (n = 125) | Controls (n = 67) | p-value |
|---|---|---|---|
| ADHD:PRS | −0.0013 (0.0002) | −0.0013 (0.0002) | 0.1817 |
| ASD:PRS | −0.0035 (0.0024) | −0.0032 (0.0021) | 0.3738 |
| BD:PRS | 0.0213 (0.0196) | 0.0211 (0.0189) | 0.9404 |
| SCZ:PRS | 0.0011 (0.0004) | 0.0011 (0.0005) | 0.7602 |
| MDD:PRS | −0.0041 (0.0018) | −0.0033 (0.0025) | 0.04754 |
ADHD:PRS: Attention-deficit hyperactivity disorder:polygenic risk score, ASD:PRS: Autism spectrum disorders:polygenic risk score, BD:PRS: Bipolar disorder:polygenic risk score, SCZ:PRS: Schizophrenia:polygenic risk score, MDD-PRS: Major depression disorder-polygenic risk score. The reported p-value is result of a Welch t-test.
SCZ and major depression shared genetic etiology with dual diagnosis
PRSs for ADHD, ASD, and BD did not share a genetic etiology at a significant level with dual diagnosis in our population: ADHD p = 0.1570, ASD p = 0.0538, and BD p = 0.1585. In contrast, SCZ and MDD:PRS each showed a significant shared genetic etiology with the dual diagnosis phenotype: SCZ (Nagelkerke Pseudo-R2 = 0.0283, corrected p = 0.0423, n = 8058 SNPs) and MDD (Nagelkerke Pseudo-R2 = 0.0451, corrected p = 0.0118, n = 334 SNPs). As can be observed, the MDD:PRS explained a higher amount of variance (4.51%) predicting placement in the dual diagnosis group than did the SCZ:PRS (2.83%).
Once we identified that MDD and SCZ PRS had a shared genetic etiology with a dual diagnosis in our sample, we performed a pair-wise comparison of the individual MDD and SCZ PRS in patients with a dual diagnosis and patients without a dual diagnosis, to determine whether these PRSs (for MDD or SCZ) could be used to detect a subgroup of patients who had SUD within each diagnostic category (BD or SCZ patients). In this analysis, patients with a dual diagnosis in the BD group had a higher MDD:PRS when compared to patients with BD without a dual diagnosis, and this difference reached statistical significance (p < 0.05) (Fig. 1). In contrast, when the MDD:PRS was applied only to patients diagnosed with SCZ, it did not distinguish between SCZ patients with and without a dual diagnosis of SUD. The SCZ:PRS did not show statistically significant differences in detecting a dual diagnosis when it was applied only to patients with SCZ (SCZ patients with and without SUD) or to patients with BD (BD patients with and without SUD). The pair-wise comparisons are shown in Table 3.
Table 3 Comparisons of MDD and SCZ polygenic risk scores
| Bipolar disorder | Schizophrenia | |||||
|---|---|---|---|---|---|---|
| Polygenic risk score | No substance use | Dual diagnosis | p-value | No substance use | Dual diagnosis | p-value |
| SCZ:PRS | 0.0010 (0.0006) | 0.0011 (0.0004) | 0.6392 | 0.0011 (0.0005) | 0.0012 (0.0004) | 0.7409 |
| MDD:PRS | −0.0048 (0.0017) | −0.0028 (0.0020) | 0.0007 | −0.0041 (0.0018) | −0.0040 (0.0016) | 0.6099 |
Dual diagnosis was considered if a patient had a psychiatric disease diagnosis (BD or SCZ) and substance use. SCZ:PRS: Schizophrenia:polygenic risk score, MDD:PRS: Major depression disorder:polygenic risk score. The reported p-value resulted from Welch t-test. BD: Bipolar disorder, SUD: Substance use disorder.
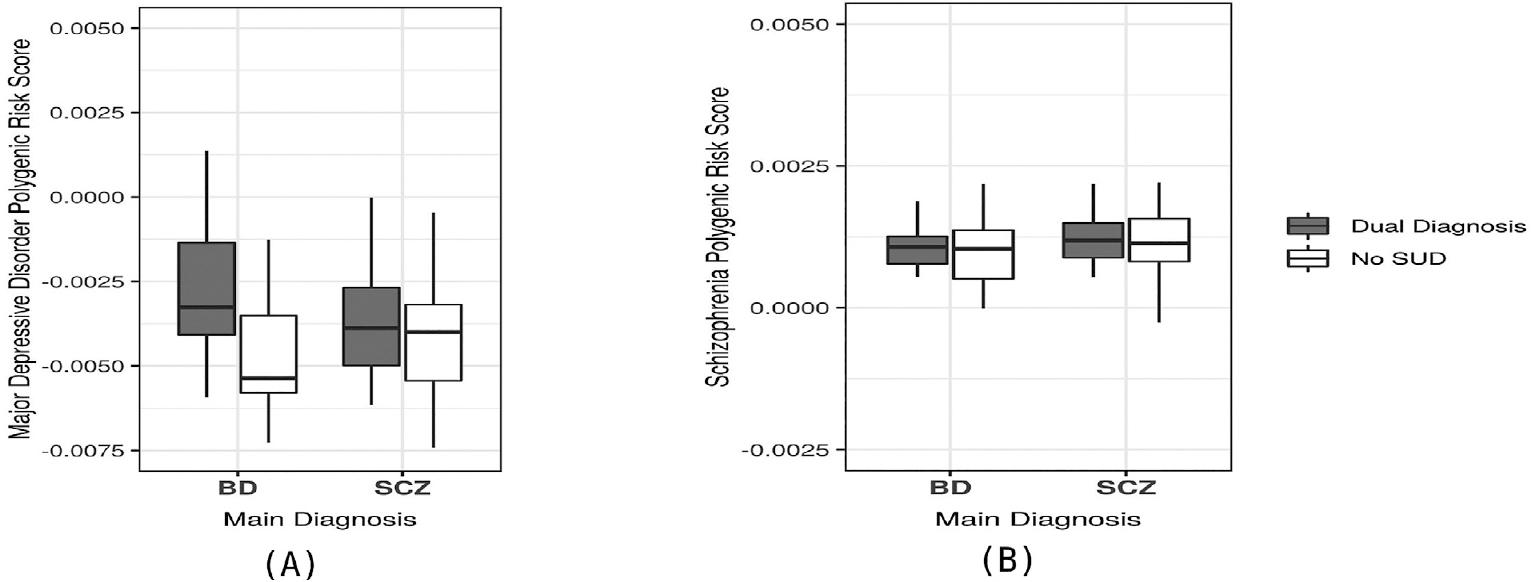
Figure 1 Comparisons of schizophrenia (SCZ) and major depressive disorder (MDD) polygenic risk score (PRS) in patients with and without dual diagnosis (substance use disorder). (A) Comparison of MDD:PRS in patients with bipolar disorder (BD) (dual diagnosis patients had significantly higher scores than BD only subjects, p = 0.0005) and patients with SCZ (non-significant difference). (B) Comparison of SCZ-PRS in BD and SCZ patients. SCZ:PRS differences between dual diagnosis patients and patients with only the primary disorder were not significantly different in either BD or SCZ subjects.
Possible global ancestry deviation of MDD and SCZ-PRS
To explore whether genetic admixture could influence PRS, we analyzed how global ancestry within the sample could affect the best-fitted PRS (also, adjusted after covariants). As PRSs of MDD and SCZ were the only ones that shared a genetic etiology with a dual diagnosis, we performed correlation tests with all the participants (SCZ, BD, and healthy controls) of each individual PRS and the global ancestry principal components (PC1 and PC2), which are the two components that separated main populations31, to search whether PRS calculations could have an ancestry-dependent deviation. In this analysis, both MDD (PC1: rho = −0.20, p = 0.01 and PC2: rho = −0.19, p = 0.01) and SCZ (PC1: rho = −0.61, p = 2.2e-16 and PC2: rho = −0.61, p = 2.2e-16) PRSs were correlated with PC1 and PC2. Of the two, SCZ:PRS had a stronger correlation with the two global ancestry components (Fig. 2).

Figure 2 Analysis of the correlation of schizophrenia (SCZ) and major depressive disorder (MDD) polygenic risk scores (PRSs) with global ancestry components. Triangles represent patients with bipolar disorder, circles represent patients with SCZ, and squares represent non-psychiatric controls. Each triangle, circle, or square represents any of the 195 individuals included in the analysis. The individuals graph in both plots is the same in the same position, the only change is the gradient of SCZ:PRS or MDD:PRS value. (A) Correlation of SCZ:PRS with global ancestry components. The gradient represents the SCZ-PRS, red color means a higher SCZ:PRS, and blue means a lower SCZ:PRS. Individuals with a higher degree of global Mexican Amerindian ancestry are grouped in values lower than zero in both principal components axes. The high correlation between the ancestry components and the SCZ:PRS is shown. (B) Correlation of MDD:PRS with global ancestry components. The gradient represents the MDD:PRS, red color means a higher MDD:PRS, and blue means a lower MDD:PRS. Individuals with a higher degree of global Mexican Amerindian ancestry are grouped in values lower than zero in both principal components axes. The low correlation between the ancestry components and the MDD:PRS is shown.
DISCUSSION
PRSs are of potential value for determining subphenotypes within a larger phenotype or main diagnosis in psychiatric disorders53,54. In the present study, we evaluated whether the current PRS (available for phenotypes of ADHD, ASD, BD, SCZ, and MDD) could also correlate with a lifetime history of dual diagnosis, in individuals diagnosed with SCZ or BD. Our results showed that both MDD and SCZ-PRSs had an impact on a dual diagnosis in the total sample. Nevertheless, when applied only to patients with one diagnosis, only MDD:PRS was found to differentiate patients diagnosed with BD and dual diagnosis from patients diagnosed with BD without a dual diagnosis.
Our study suggests that both the MDD:PRS and the SCZ:PRS might be of use in detecting risk for a dual diagnosis; however, when PRSs are applied only to a specific diagnosis, we suggest that MDD:PRS, used in patients with BD, is the only specific PRS which correlates with a dual diagnosis within the specific diagnoses of SCZ or BD, respectively. The shared genetic susceptibility between MDD and alcohol dependence (AD) might be what drove this result within our BD patients, especially when noting that the main substance of SUD in our samples was (apart from nicotine) alcohol abuse/dependence. These findings are consistent with the study of Andersen et al.55, where they suggested that shared genetic susceptibility contributed to MDD and AD comorbidity.
Although other studies have applied PRS to explore the shared genetic background between psychiatric disorders and SUDs24,25, all the approximations have been made in populations of European ancestry and not in admixed populations. Our study is one of the first approximations on how to apply psychiatric PRS in admixed populations. Our results suggest that PRS must be applied with caution in admixed populations such as the Mexican population, which has individuals with varying levels of admixture31,49,56. In relation to this, we found that SCZ:PRS showed a correlation with global ancestry components. The difference in PRS based on demographic history has been previously explored32,57. Martin et al. evaluated eight complex traits PRS in the 1000 Genomes Project panel and found similar results to ours; they observed that SCZ:PRS could be deviated based on the main populations ancestry. In their analysis, they also reported that it was not possible to predict how PRS could change according to ancestry. In this sense, we think that the application of PRS in different populations, with distinct admixtures and diverse phenotypes, could give us more information on the use of PRS for psychiatric disorders as a translational risk prediction tool.
The results obtained from this study should be considered as preliminary due to the small sample size; it will be necessary to increase the sample size to have a better understanding of both the utility of PRS to determine dual diagnosis risk in BD and SCZ, and to assess how to correct for genetic population factors that influence PRS. This study is among the first ones looking at how PRSs for psychiatric disorders perform as markers of dual diagnosis in admixed populations. Nevertheless, we found that dual diagnosis had a shared etiology with MDD and SCZ. The present study can help reduce disparities in what is known about the PRSs in different populations.











 text new page (beta)
text new page (beta)


