Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista de investigación clínica
versão On-line ISSN 2564-8896versão impressa ISSN 0034-8376
Rev. invest. clín. vol.58 no.4 Ciudad de México Jul./Ago. 2006
Carta al editor
Confirmation of tyrosine 698 in beta subunit of cGMP phosphodiesterase in patients with retinitis pigmentosa and population of the west of Mexico
Isabel Cristina Núñez–Gutiérrez,* ** Diana García–Cruz,* Rubén Fragoso–Herrera,* Claudina Medina–Lozano*
* División de Genética, Centro de Investigación Biomédica de Occidente, Instituto Mexicano del Seguro Social de Guadalajara.
** Genética Humana, Centro Universitario de Ciencias de la Salud, Universidad de Guadalajara.
Correspondence and reprint request:
Isabel Cristina Núñez–Gutiérrez,
División de Genética, Centro de Investigación Biomédica de Occidente, IMSS.
Sierra Mojada 800, Col. Independencia,
44340, Guadalajara, Jal.
Phone: (52–33) 36189410, fax: (52–33) 36181756.
E–mail: isabelcristinanuñez@yahoo.com.mx
Recibido el 13 de enero de 2006.
Aceptado el 13 de junio de 2006.
INTRODUCTION
The cGMP phosphodiesterase beta–subunit (PDEB), when associated with a cGMP phosphodiesterase alpha–subunit, forms a functional holoenzyme that participates in the processes of transmission and amplification of the visual signal in rod cells. Both subunits have two cGMP–binding sites (GAF) in the N–terminal half and a catalytic domain (PDEase) in the C–terminal portion.1 The PDEB gene (4pl6.3) presents with at least 28 reported mutations causing hereditary retinitis pigmentosa (RP), 27 polymorphic sites, and seven regions in conflict.1 'Regions in conflict' are those with more than one version of the nucleotide sequence encoded by a specific DNA region. The conflict at site 698 in the PDEB protein is caused by the putative assignment of the amino acids isoleucine (He) or tyrosine (Tyr), coded by ATC and TAC triplets, respectively. Both triplets differ in the two first nucleotides (AT or TA), corresponding to positions 71 and 72 of exon 17, located in the PDEase domain of PDEB.1 Weber and co–workers first reported the complete sequence of PDEB as having AT in region 698, but then Collins and co–workers reported TA for the same region.1 Both studies were performed in clones of genomic DNA or cDNA, respectively. To date, any one of the seven sites in conflict of the human PDEB have been resolved. We carried out a survey to resolve the conflict site 698, by studying the nucleotides sequence of human genome samples directly. We analyzed PCR products of exon 17 of one hundred Mexican individuals for recognize TAC triplet by restriction enzyme assay and by sequencing.
MATERIAL AND METHODS
Subjects
Fifty affected individuals with hereditary RP and 50 healthy individuals from the west of Mexico were studied.
Methods
DNA from whole blood was extracted using a salting–out procedure. Polymerase chain reaction (PCR) amplification of exon 17 and a restriction enzyme assay were used for identification of triplet TAC. The Rsal restriction enzyme recognizes and cuts the unique 5'–GT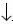 AC–3' sequence, contained in the triplet coding for Tyr. PCR amplification was performed under standard conditions with an annealing temperature of 62 °C, using the forward primer 5'–GGG GCG GGG TCT CCA CAC TT–3' and the reverse primer 5'–GGA CCC CTC GTG CCC CCT CC–3'.2 PCR products were digested withñsal (New England Biolabs), electrophoresed in 12% polyacrylamide gels, and stained with 0.2% silver nitrate solution. The PCR products of exon 17 were amplified on both senses from two patients with RP and from one healthy individual. They were sequenced using ABI PRISMTM BigDye Terminator Chemistry on an Applied BioSystems 3100 Sequencer (Applied BioSystems, Foster City, CA, USA).
AC–3' sequence, contained in the triplet coding for Tyr. PCR amplification was performed under standard conditions with an annealing temperature of 62 °C, using the forward primer 5'–GGG GCG GGG TCT CCA CAC TT–3' and the reverse primer 5'–GGA CCC CTC GTG CCC CCT CC–3'.2 PCR products were digested withñsal (New England Biolabs), electrophoresed in 12% polyacrylamide gels, and stained with 0.2% silver nitrate solution. The PCR products of exon 17 were amplified on both senses from two patients with RP and from one healthy individual. They were sequenced using ABI PRISMTM BigDye Terminator Chemistry on an Applied BioSystems 3100 Sequencer (Applied BioSystems, Foster City, CA, USA).
RESULTS
All 100 samples were positive for the Rsal restriction site (Figure 1). The TAC nucleotides were present at positions 107–109 of the electropherograms of all three analyzed samples (Figure 2). There were no samples with partial Rsal digestion, nor did the electropherograms show any double signals at these positions, discounting the presence of a heterozygous state. Thus, the TAC triplet was present in all 200 chromosomes and, consequently, Tyr is located at the putative conflict site 698 of all individuals.


DISCUSSION AND CONCLUSION
The PDEase domain is present in a superfamily of enzymes with phosphohydrolase activity involved in the phototransduction signal, nucleic acid metabolism and other functions. This catalytic domain is widely distributed among plants, invertebrates and mammals.3 Except for the sequence reported by Weber and co–workers, all the PDEB clones from humans and other mammalian species show the presence of TA nucleotides at the putative conflict region 698 (Blastn 2.2.5, RID: 1048776668–018755–28686).4
Three–dimensional analysis of the human PDEB protein indicates that Tyr698 occupies a position with a prominent external projection (Swiss–Model, jobid: AAAaOM_JZ).5 A hypothetical replacement of Tyr698 (a polar amino acid) by He (a non–polar amino acid) produce an important alteration in the structural conformation of the PDEase domain and would probably also affect the enzymatic activity of the PDEB protein. Although there are no reported mutations at position 698 leading to visual defects, the adjacent mutation Leu699Arg produces hereditary RP;1 therefore, we hypothesize that Tyr698 plays a major role in the PDEase domain.
Our results show that all 200 chromosomes analyzed have the TAC triplet and that presence of Tyr698 in the PDEB protein does not alter the function of the phosphodiesterase involved in the visual phototransduction signal because there was no difference between patients with RP and healthy individuals. Together, it is reasonable to conclude that the site 698 should no longer be considered a region in conflict on the human PDEB appointment, and on the other hand, it is open the possibility that ATC triplet could to be product of the management biotechnological of the clone, or, it represents a mutant or polymorphic allele.
ACKNOWLEDGMENTS
This work was supported by the grant number FP2001/048 from Fondo para el Fomento de la Investigación (FOFOI), Instituto Mexicano del Seguro Social. The authors wish to thank Dr. Maria Isabel Hernández Lugo, head of Banco de Sangre Central of Centro Médico de Occidente–IMSS, for donating the blood samples from volunteer donors.
REFERENCES
1. The Universal Protein Resource (UniProt) [database on the Internet] in Protein Information Resource. UniProtKB Entry: P35913 [last annotations in 2006–04–18]. Available from: http://www.pir.uniprot.org/cgi–bin/upEntry?id=P35913. [ Links ]
2. McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the P subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Nati Acad Sci USA 1995; 92: 3249–53. [ Links ]
3. Interpro [database in Internet] in European Molecular Biology Laboratory/ European Bioinformatics Institute EMBL/EBI. ID: IPR002073. Available from: http://www.ebi.ac.uk/interpro/IEntry?ac=IPR002073 [ Links ]
4. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402. [ Links ]
5. Peitsch MC. Protein modelling by E–mail. Bio/Technology 1995; 13: 658–660; Guex N, Peitsch MC. Swiss–Model and the Swiss–PdbViewer. An environment for comparative protein modelling. Electrophoresis 1997; 18: 2714–2723; [ Links ]
6. Schwede T, Kopp J, Guex N, Peitsch MC. Swiss–Model: an automated protein homology–modeling server. Nucleic Acids Res 2003; 31: 3381–5. [ Links ]














