Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Revista de investigación clínica
versão On-line ISSN 2564-8896versão impressa ISSN 0034-8376
Rev. invest. clín. vol.56 no.1 Ciudad de México Fev. 2004
Rev Invest Clín 2004; Vol. 56(1):43-50
ARTÍCULO ORIGINAL
Production of recombinant rat hepatic histidase
Producción de histidasa recombinante de hígado de rata
Victor Ortiz
Nimbre Torres
Armando R. Tovar
Depto. Fisiología de la Nutrición, Instituto Nacional de Ciencias Mèdicas y Nutrición Salvador Zubirán, Mèxico, D.F.
Correspondence and reprint request:
Armando R. Tovar M.D.
Departamento de Fisiología de la Nutrición
Área de Nutriología Molecular
Instituto Nacional de Ciencias Médicas y Nutrición
Salvador Zubirán Vasco de Quiroga Nª 15
14000 México D.F.
Tel:5573-1200 ext 2802 o 5655-3038;
Fax: 5655-1076
E-mail artovar@quetzal.innsz.mx
Recibido el 25 de marzo de 2003
Aceptado el 23 de septiembre de 2003
ABSTRACT
Rat liver histidase was expressed in E. coli by using a PCR product of the coding sequence obtained from the rat liver cDNA of histidase cloned in the expression vector pRSET. The construct (pRSET-HAL) produced a fusion protein containing a tail of polyhistidine. The expression product was purified with a resin containing Ni+ that retains proteins with polyhistidine fragments. pRSET-HAL was analyzed by restriction enzyme mapping and by sequencing confirming the correct orientation and nucleotide sequence. Native rat liver histidase was also purified and it had a Mr of 72 kDa. An antiserum against native histidase was obtained in rabbit. Western blot analysis revealed one band of 72 kDa observed in membranes containing purified histidase or rat liver high speed supernatants. The same antiserum also detected in cell lysates of E. coli transformed with the pRSET-HAL plasmid a single band of 74 kDa of the recombinant histidase before cleavage with enterokinase. After the proteolysis, the Western blot analysis showed a single band of approximately 72 kDa. Kinetic analysis of the recombinant histidase showed similar Km and Vmax compared with native histidase.
KEY WORDS. Histidase. Liver. Rat.
RESUMEN
La histidasa de hígado de rata se expresó a partir del producto de amplificación por PCR de la región codificante del DNAc de la histidasa, el cual se clonó en el vector de expresión pRSET. La construcción (pRSET-HAL) permitió la producción de una proteína de fusión que contenía una cola de polihistidinas. El producto de expresión se purificó con una resina que contiene Ni+, la cual retiene proteínas con fragmentos de polihistidina. El pRSET-HAL se analizó por medio de un análisis con enzimas de restricción y por secuenciación de DNA para confirmar su correcta orientación así como tambièn su secuencia nucleotídica. La histidasa nativa hepática fue tambièn purificada y tuvo un Mr de 72 kDa. A partir de la enzima purificada nativa se preparó un antisuero contra la histidasa en conejo. El análisis por Western blot mostró una sola banda de 72 kDa en membranas que contenían histidasa purificada o en la fracción citosólica de hígado de rata. El mismo anticuerpo tambièn detectó en lisados de E. coli transformados con el plásmido pRSET-HAL una sola banda de 74 kDa correspondientes a la histidasa recombinante antes de su hidrólisis con enterocinasa. Despuès de la proteólisis, el análisis por Western blot mostró una sola banda de aproximadamente 72 kDa correspondiente a la histidasa. Análisis cinèticos de la histidasa recombinante mostraron Km y Vmax similares a las observadas con la histidasa nativa.
PALABRAS CLAVE. Histidasa. Hígado. Rata.
INTRODUCTION
Histidase (L-histidine ammonia-lyase, E.C.4.3.1.3) is the first enzyme in the degradation of histidine, producing urocanic acid. 1,2 Histidase is expressed only in liver and epidermis. 3,4 In the liver, the trans-urocanic acid is metabolized to glutamate and formimino tetrahydrofolate, the latter an important compound in the one-carbon pool metabolism. However, urocanase is not naturally present in the skin, and consequently, urocanic acid accumulates in the epidermis. Urocanic acid protects DNA from photomutagenesis 5 and may act as a mediator of the systemic immunosuppresion of tumor rejection. 6 Histidase, a cytosolic enzyme, contains a dehydroalanine residue in its active site essential for the deamination of histidine. 7 The rat histidase cDNA has been cloned, and it has an open reading frame of 1,971 bp coding for a polypeptide of 657 amino acids with a Mr of 72,165 Da. 3 The human histidase gene has been sequenced, and it is a single copy gene of 25 Kb containing 21 exons. 8,9 The deficiency of histidase results in histidinemia, which is characterized by increased plasma histidine and histamine, reduced urocanic acid, and increased histidine metabolites such as imidazole pyruvic acid, the direct product of histidine transamination. 10 However, it is considered a metabolic benign disease. 11,12 Hepatic histidase gene expression, as well as most of the amino acid degrading enzymes, is increased by the ingestion of high protein diets 4,13,14 or by the administration of hormones such as glucagon or glucocorticoids. 4,15 The only amino acid degrading enzyme unaffected by the protein content of the diet 14 or by hormones 15 is the branched-chain aminotransferase, which is induced only under conditions of rapid cell duplication. 16,17 Histidase expression is also increased in catabolic states such as nephrotic syndrome, 18 or by consuming histidine imbalanced diets. 19 During severe malnutrition in rats, the expression of this enzyme is repressed, and after protein repletion, mRNA concentration responds rapidly to the concentration of dietary protein 20 in parallel with some indicators of the nutritional status such as IGF-1. 21 In most of the previous studies, the quantitation of the amount of histidase has been done by Western blot analysis. However, the purification of rat liver histidase for preparing the antibody against this enzyme is time consuming. Thus, the purpose of this study was to produce recombinant histidase in a bacterial expression system and to measure its immunoreactivity with antibodies raised against the native hepatic histidase as well as its kinetic properties.
MATERIALS AND METHODS
Purification of native liver histidase
1. Sample preparation. Livers from 4 female Wistar rats weighing between 250 to 300 g were obtained, and immediately washed in cold saline solution and blotted. Livers were weighed and 3 mL of 10 mM Tris-HCl, pH 7.4, containing 14 mM MgCl 2 and 0.6 mM KCl were added per each gram of tissue. Livers were homogenized with a Polytron instrument (Kinematica, Switzerland) at the lowest setting, the homogenate was centrifuged at 92,500 g for 50 min at 4 °C in an ultracentrifuge (XL-90; Beckman, Palo Alto, CA). The clear supernatant was carefully collected, transferred to a beaker and heated to 65 °C in a water bath for 5 min. The sample was then centrifuged at 47,000 g for 15 min at 4 °C (J2-MC; Beckman, Palo Alto, CA) and the precipitate was discarded.
2. Ammonium sulfate fractionation.
Ammonium sulfate was added to the above supernatant while it was being stirred to reach a 38% saturation (228.6 g/L). The mixture was stirred for 30 min at 4 °C and centrifuged at 47,000 g for 20 min at 4 °C. The precipitate was discarded and the supernatant was again fractionated with the addition of ammonium sulfate to reach a 50% saturation (75.2 g/L). The mixture was stirred for 30 min at 4 °C and centrifuged at 47,000 g for 20 min at 4 °C. The precipitate was dissolved in a minimum volume of cold 0.1 M Tris-HCl, pH 7.6
3. Chromatography on DEAE-Sephadex .
The resuspended precipitate of the previous step was applied to a column of 2 x 30 cm containing DEAE-Sephadex equilibrated with 0.1 M Tris-HCl, pH 7.6, containing 0.1 M NaCl. DEAE-Sephadex was prepared by mixing 1 g of the resin with the equilibration buffer and then deaerating for 20 min. The sample was eluted with the same buffer, and fractions of 4.4 mL were collected and dialyzed against 4 L of 20 mM Tris-HCl, pH 7.6, overnight at 4 °C. Fractions were assayed for histidase activity; active fractions were pooled and 2 mg/mL of DTT was added to maintain the enzyme activity.
4. Ammonium sulfate fractionation .
The pooled fractions from the previous column was precipitated by adding ammonium sulfate to reach 40% saturation (242.3 g/L). The mixture was stirred for 15 min at 4 °C and centrifuged at 40,000 g for 10 min at 4 °C. The supernatant was then fractionated again with ammonium sulfate to reach a 50% saturation (62.7 g/L). The mixture was stirred for 15 min at 4 °C, transferred to tubes and centrifuged at 40,000 g for 10 min at 4 °C. The precipitate was dissolved in a minimum volume of cold 0.1 M imidazole chloride, pH 6.8.
5. Chromatography on QAE-Sephadex.
The resuspended precipitate of the previous step was applied to a column of 1.5 x 20 cm containing QAE-Sephadex equilibrated with 0.1 M imidazole chloride, pH 6.8. QAE-Sephadex was prepared by mixing 1 g of the resin with 25 mL of 0.1 M imidazole chloride pH 6.8 and deacreated for 20 min. Samples were eluted with a linear gradient from 0 to 0.5 M NaCl dissolved in 0.1 M imidazole chloride, pH 6.8, and fractions of 2 mL were collected. Fractions were assayed for histidase activity and active fractions were pooled.
6. Sample concentration.
The pooled fractions were concentrated with an Amicon Centricon 50 device according to the manufacturer instructions. The protein was stored at -20 °C for no more than two weeks since histidase was unstable and began to degrade.
Protein assay and enzyme purity
Protein concentration was measured using the Bio Rad assay, based on the Bradford dye-binding procedure. Enzyme purity was assessed by SDS/PAGE using the method of Laemmli with gels containing 7.5% (w/v) polyacrylamide. Gels were stained with Coomassie Blue. The molecular weight of histidase was determined with reference to low molecular weight markers (Bio Rad; Hercules, CA).
Preparation of antiserum against rat liver histidase
Purified homogenous histidase (200 µg) was emulsified in an equal volume of complete Freund's adjuvant. This suspension was injected subcutaneously into a rabbit and was followed by a second injection two weeks after the first with the same concentration of protein mixed with incomplete Freund's adjuvant; one booster injection was applied two weeks later. Blood was collected two weeks after the last immunization. IgG fraction was partially purified.
Immunoglobulin fraction purification
Thirty mL of serum containing anti-histidase antibodies were fractionated with 60 mL of 33% (w/v) ammonium sulfate solution. The mixture was stirred overnight at 4 °C and centrifuged at 12,000 g for 20 min at 4 °C. The precipitate was resuspended in 30 mL of 33% (w/v) ammonium sulfate solution and centrifuged again as above, and washed again with the same ammonium sulfate solution. The precipitate was resuspended in 3 mL of 0.1 M phosphate-buffered saline (PBS) containing 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na 2 HPO 4 , 1.4 mM KH 2 PO 4 , pH 7.2, dialyzed 48 h at 4 °C with the same buffer. The antiserum was kept at -20 °C in aliquots of 50 μL.
Western-blot analysis and neutralization of histidase activity with the antiserum
A sample of 50 μg pure histidase or rat liver cytosolic supernatant were separated on 7.5% (w/v) polyacrilamide gel by SDS/PAGE and transferred to nitrocellulose (Biorad) as described by Towin. 22 Membranes were incubated for 2 h at room temperature in 10% (w/v) skim milk in PBS/Tween buffer and washed three times with PBS/Tween. The membrane was incubated 2 h with histidase antiserum diluted 1:50 in PBS/Tween, washed three times with PBS/Tween, and incubated for 2 h with goat antirabbit IgG gamma-chain specific affinity-purified antibody peroxidase conjugate (KPL, Inc.) diluted 1:1,000 in PBS/Tween. Membranes were washed five times with PBS/Tween. Immunoreactive protein band was visualized after horseradish peroxidase reduction of 4-chloro-1-naphtol. As negative control preinmune serum was used. Neutralization of histidase activity with a sample of 400 μL of cytosolic supernatant were mixed with different amounts of antiserum and incubated at 4 °C for 90 min, and then centrifuged at 14,000 g for 10 min at 4 °C. The supernatants were assayed for histidase activity.
Histidase assay and kinetic measurements
Histidase activity was assayed according to Tabor and Mehler 23 in 10 mM sodium pyrophosphate pH 9.2. Two hundred µL of the sample was added to the buffer, and the reaction was initiated by the addition of 1 mL of 0.1 M L-histidine, pH 9.2. The final reaction volume was 3 mL. Assays were carried out at 25 °C for 10 min, and the rate of urocanic acid produced was monitored spectrophotometrically at 277 nm. A histidase unit was defined as 1 nmol of urocanic acid produced per minute at 25 °C. Values for Km and Vmax were determined using the enzyme mechanism mode included in a DU640 spectrophotometer (Beckman, Palo Alto CA, USA) from a set of samples (0.36 to 10 mM substrate concentration) using non-linear regression analysis.
Expression and purification of recombinant histidase
1. Plasmids and PCR.cDNA for rat liver histidase (pHAL) was kindly provided by Dr. Roederick McIness. 3 The cDNA of 2.18 Kb was cloned into the EcoR I site of Bluescript KS(-). The coding sequence was amplified by PCR by using two oligonucleotides where unique restriction sites were introduced to create an in-frame fusion protein. The upper primer was a 28-mer GCGGGATCCATGCCTAGGTACACGGTGC and the lower primer was a 29-mer GCGAAGCTTTTAAAGATCGTCAGACTCTG. Primers were synthesized with an Oligo 1,000 DNA synthesizer (Beckman, Palo Alto, CA, USA). One ng of pHAL, 5 μL of 10 µM each primer, 0.2 M dNTPs, 1 X Taq buffer and 2.5 units of Taq polymerase were combined in a reaction volume of 100 μL. The reaction conditions were 94 °C for 1 min followed by 25 cycles, each consisting of 1 min at 94 °C, 1 min at 52.5 °C, 1.10 min at 72 °C, followed by a single cycle extension for 7 min at 72 °C. The product of 1,971 bp was purified with Geneclean (Bio 101, Vista CA, USA), treated with BamH I and Hind III and ligated in the Bam H I-Hind III cloning site of the pRSET vector which expresses polyhistidine-containing recombinant proteins (Invitrogen; San Diego, CA, USA). E. coli strain used for expression was JM109. The PCR product cloned in pRSET was sequenced 24 to verify that there were not changes in the nucleotide sequence of the amplified product.
2. Expression and purification of the fusion protein.
Two mL of SOB medium (20 g tryptone, 5 g yeast extract, 0.5 g NaCl, 186 mg KCl in 1 L of deionized water, pH 7.0 containing 10 mM MgCl 2 2)containing ampicillin (50 μg/mL) were inoculated with a single recombinant E. coli colony and grown overnight at 37 °C with shaking. Fifty mL of SOB medium containing ampicillin (50 μg/mL) were inoculated with 0.3 mL of the overnight culture and grown at 37 °C to an A 600 l 0.3. Expression of the fusion protein was induced by the addition of isopropyl a-D-thiogalactoside (IPTG) at a final concentration of 1 mM. Cells of E. coli were grown for an additional hour, and then were infected with M13/T7 phage (5 plaque-forming units/cell) that was previously titrated. Culture was grown at 37 °C for an additional 4 h, pelleted by centrifugation at 4,000 g for 10 min at 4 °C. Purification of the protein was carried out with a high affinity ProBond resin according to the manufacturer's instructions (Invitrogen, San Diego, CA, USA); ProBond resin is able to bind fusion protein containing several residues of histidine in the N-terminal of the protein, such as the fusion proteins expressed with pRSET vector. Recombinant histidase was cleaved with enterokinase to remove the polyhistidine residue. Proteins were analyzed with SDS-PAGE gels and specificity was determined with Western blot.
Statistics
Differences in the kinetic constants between recombinant histidase and native histidase were assessed by unpaired t-test using the Statview program (Abacus Concepts, Inc., Berkeley, CA) for the Macintosh computer. Differences were considered significant at P <0.05.
RESULTS AND DISCUSSION
Purification of rat liver histidase Table 1 shows the purification of rat liver histidase. The purification protocol involved five steps. Heating of the supernatant at 65 °C increased the specific activity 4-fold. The first ammonium sulfate fractionation produced a red precipitate which contained histidase activity, and there was an increase of the specific activity by 24-fold in comparison with the homogenate. The next fractionation step was carried out in a DEAE-Sephadex column equilibrated with 0.1 M Tris-HCl, pH 7.6, containing 0.1 M NaCl
(Figure 1A); proteins were eluted with the same buffer and almost all histidase activity was collected in fractions 4 and 5, increasing the specific activity by 44-fold. The second ammonium sulfate fractionation, which produced a white precipitate containing histidase activity, increased by 1.1-fold the specific activity in comparison with the previous step of purification. The precipitated protein was resuspended in 0.1 M imidazole chloride, pH 6.8, and it was applied onto a QAE-Sephadex column (Figure 1B) resulting in a 4.3-fold increase in the specific activity with respect to the previous step and an overall 500.3-fold total purification of the enzyme with a final yield of about 0.65 % (Table 1). SDS-PAGE of the different steps of the purification of histidase is shown in (Figure 2). After the chromatography with QAE-Sephadex, the gel showed one band, corresponding to pure histidase. By comparison with molecular weight standards ranging from 97.4 to 14.4 kDa, a molecular mass value of 71.8 kDa was calculated for the pure protein. Mammalian histidase measured in native gels has a molecular mass of about 200 kDa and it has been reported that it is formed by three identical units of approximately 75 kDa each. 25,26 Cloned rat liver histidase cDNA, detects a single mRNA of 2.5 kb in rat liver and skin. 3,4 The cDNA has an open reading frame of 1,971 bp coding for a polypeptide of 657 amino acids with a molecular mass of 72,165 Da, 3 confirming the molecular mass obtained for a subunit of histidase in this study. Similarly, the human histidine gene codes for a protein of 72,651 Da, and its amino acid sequence has a similarity of 93% with the mice and rat sequences. 8 In contrast, bacterial histidase contains subunits of approximately 55 kDa organized in tetramers. 27
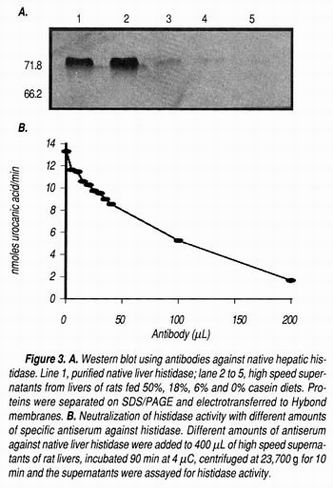
The antiserum against histidase was specific for this enzyme and recognized only a band of 71.8 kDa in a Western blot analysis (Figure 3A). This antiserum was able to neutralize almost 90% of histidase activity in liver high speed supernatants (Figure 3B), showing the specificity of the antiserum. Preimmune serum was not able to detect any band in the blot. The stability of the purified enzyme was only 3 weeks, and after this period, several bands in a SDS-PAGE appeared with a molecular weight below 50,000. It has been reported that degradation of Streptomyces griseus histidase showed the appearance of polypeptides derived from the hydrolysis between amino acids 66 and 67 of the protein. 27
Expression of recombinant histidase in E. coli
The cDNA containing the coding sequence of rat histidase was amplified by PCR. The product obtained showed a size of 1,971 bp that included the complete coding sequence. The size of the full length cDNA is 2,179 bp. The PCR product contained a 5' Bam H I restriction site and a 3- Hind III site, which in turn was ligated in the Bam H I-Hind III cloning site of the expression vector pRSET. Restriction analysis using Bam H I and Hind III as well as DNA sequencing were performed to verify that the PCR product was in the correct orientation for its expression. No errors were introduced in the nucleotide sequence of the PCR product cloned in pRSET. The resulting expression plasmid (pRSET-HAL) produced a fusion protein with a polyhistidine residue and a site of enterokinase cleavage recognition sequence. Induction of E. coli JM109 containing pRSET-HAL with isopropyl β-D-thiogalactopyranoside (IPTG) showed the presence of inclusion bodies after 4 h of incubation only when cultures were incubated at 35 °C. Incubation at 36 or 37 °C did not stimulate cells to synthesize the protein. SDS-PAGE analysis of the bacterial lysate during the 5 hours of induction showed an increased expression of a protein of 74 kDa which corresponded to the molecular weight of a protein that included the hepatic histidase, the polyhistidine tail, and the enterokinase protease site (Figure 4A). Immunoblot analysis of the bacterial lysate after 5 hours of induction using anti-histidase antiserum detected a single band of approximately 74 kDa (Figure 4B, line 1). The overexpressed protein after 5 hours of induction was purified by using a Probond resin (Invitrogen, San Diego, CA). Digestion with enterokinase produced a protein of approximately 72 kDa, corresponding to the molecular mass of native hepatic histidase (Figure 4B, lane 2). The purified protein showed histidase enzyme activity, with a specific activity similar to the observed with the native protein. It has been reported that the presence of a residue of dehydroalanine derived from a serine located in the position 254 of the amino acid sequence of histidase is essential for the activity of this enzyme, and this modified residue was apparently conserved since the overexpressed histidase maintained the enzyme activity. The kinetic data of purified native histidase showed a Km of 1.52 ±0.43 mM for histidine with a Vmax of 1.68 ± 0.17 nmoles urocanic acid/min per mg of protein (Figure 4C). The Km value is close to that reported by Okamura of 1.2 mM. 25 This enzyme was not inhibited by its product, urocanic acid (results not shown). On the other hand, the Km for the recombinant histidase was 1.75 ± 0.54 mM with a Vmax of 1.92 ± 0.22 nmoles urocanic acid/min per mg of protein. Despite the changes observed in Km and Vmax values for the native and the recombinant enzymes, there were no significant differences between them.
Isolation of native histidase is a time consuming procedure, and the isolated protein is stable for only two weeks. Production of recombinant histidase reduces time and increases the amount of protein isolated, facilitating the use of the protein for different purposes such as production of antibodies against histidase as well as the structural analysis of this enzyme. With this tool we will extend our studies on the regulation of hepatic histidase. We have clear evidence that the mRNA of this enzyme increases when rats are fed with a high protein diet, or by the injection of hormones such as glucagon or glucocorticoids. 4,15 The increase in histidase mRNA abundance is associated with an increase in histidase gene transcription. We have evidence by Western blot analysis, using antibodies against the native histidase, that the increment of mRNA concentration shows a parallel increment with the amount of histidase protein. 4 However, it is not know what type of liver cells synthesize histidase, and if there is a zonal distribution of this enzyme between periportal and perivenous hepatocytes, as has been observed for other enzymes. 28-30 With the production of the antibodies against recombinant histidase we will be able to study the liver distribution of histidase. In conclusion, the results of this study showed that overexpressed recombinant histidase with a Mr of approximately 72 kDa, was recognized by a specific antibody risen against native purified rat liver histidase. The enzyme maintained its biological activity, and it can be used as a tool for studying regulation of histidine metabolism, histidase tissue distribution, and its structural analysis.
ABBREVIATIONS
●BSA: bovine serum albumin.
● DTT: DL-dithiothreitol.
● DEAE: diethylaminoethyl.
● IPTG: isopropyl β-D-1-thiogalactopyranoside.
● dNTPs: deoxynucleotides.
● pRSET: expression vector derived from the pUC vector.
● pRSET-HAL: pRSET vector containing the coding sequence of histidase.
● PBS: phosphate-buffered saline.
● pHAL: cDNA sequence cloned in p-Bluescript vector.
● PCR: polymerase chain reaction.
● QAE: diethyl-(2-hydroxypropyl) aminoethyl.
● SDS-PAGE: sodium dodecyl sulfate-polyacrylamide gel electrophoresis
● SOB: Hanahan's broth.
REFERENCES
1. Mehler AH, Tabor H. Deamination of histidine to form urocanic acid in liver. J Biol Chem 1953; 201: 775-84. [ Links ]
2. Peterkofsky A. The mechanism of action of histidase: Amino-enzyme formation and partial reactions. J Biol Chem 1962; 237: 787-95. [ Links ]
3. Taylor RG, Lambert MA, Sexsmith E, Sadler SJ, Ray PN, Mahuran DJ, McInnes RR. Cloning and expression of rat histidase. J Biol Chem 1990; 265: 18192-9. [ Links ]
4. Torres N, Martínez L, Alemán G, Bourges H, Tovar AR. Histidase expression is regulated by dietary protein at the pretranslational level in rat liver. J Nutr 1998; 128: 818-24. [ Links ]
5. Morrison H, Avnir D, Azrella T. Z/E photoisomerization of urocanic acid. Photochem Photobiol 1980 a; 32: 711-4. [ Links ]
6. De Fabo EC, Noonan FP. Further evidence that the photoreceptor mediating UV-induced systemic immune suppression is urocanic acid . J Invest Dermatol 1983; 80: 319. [ Links ]
7. Taylor RG, McInnes RR. Site-directed mutagenesis of conserved serines in rat histidase. J Biol Chem 1994; 269: 27473-7. [ Links ]
8. Suchi M, Harada N, Wada Y, Takagi Y. Molecular cloning of a cDNA encoding human histidase. Biochim Biophys Acta 1993; 1216: 293-5. [ Links ]
9. Suchi M, Sano H, Mizuno H, Wada Y. Molecular cloning and structural characterization of the human histidase gene (HAL). Genomics 1995; 29: 98-104. [ Links ]
10. La Du BN. L-histidine ammonia-lyase (Human stratum corneum). Meth Enzymol 1971; 17B: 891-7. [ Links ]
11. Taylor RG, Levy HL, McInnes RR. Histidase and histidinemia. Clinical and molecular considerations. Mol Biol Med 1991; 8: 101-16. [ Links ]
12. Virmani K, Widhalm K. Histidinemia: A biochemical variant or a disease. J Am Coll Nutr 1993; 12: 115-24. [ Links ]
13. Rodríguez B, Torres N, Rincon AR, Bourges H, Panduro A, Tovar AR. Hepatic phenylalanine hydroxylase and tyrosine aminotransferase mRNA levels in rats adapted to diets with different concentrations of protein. Rev Invest Clin 1996; 48: 413-9. [ Links ]
14. Torres N, López G, DeSantiago S, Hutson SM, Tovar AR. Dietary protein level regulates expression of the mitochondrial branched-chain aminotransferase in rats. J Nutr 1998; 128: 1368-75. [ Links ]
15. Alemán G, Torres N, Bourges H, Tovar A. Regulation of histidase gene expression by glucagon, hydrocortisone and protein-free/high carbohydrate diet in the rat. Life Sci 1998; 63: 1663-73. [ Links ]
16. Tovar AR, Becerril E, Hernández-Pando R, López G, Suryawan A, DeSantiago S, Hutson SM, Torres N. Localization and expression of BCAT during pregnancy and lactation in the rat mammary gland. Am J Physiol Endocrinol Metab 2001; 280: E480-E8. [ Links ]
17. Torres N, Vargas C, Hernández-Pando R, Orozco H, Hutson SM, Tovar AR. Ontogeny and subcellular localization of rat liver mitochondrial branched chain amino-acid aminotransferase. Eur J Biochem 2001; 268: In press. [ Links ]
18. Ascencio C, Tovar AR, Medina-Campo ON, Pedraza-Chaverri J, Torres N. Hepatic histidase and muscle branched chain aminotransferase gene expression in experimental nephrosis. Life Sci 2000; 67: 2775-84. [ Links ]
19. Torres N, Berinstain L, Bourges H, Tovar AR. Histidine imbalanced diets stimulate hepatic histidase gene expression in rat. J Nutr 1999; In press. [ Links ]
20. Tovar AR, Santos A, Halhali A, Bourges H, Torres N. Hepatic histidase gene expression responds to protein rehabilitation in undernourished growing rats. J Nutr 1998; 128: 1631-5. [ Links ]
21. Tovar AR, Halhali A, Torres N. Effect of nutritional rehabilitation of undernourished rats on serum insulin-like growth factor (IGF)-1 and IGF-binding proteins. Rev Invest Clin 1999; 51: 99-106. [ Links ]
22. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA 1979; 76: 4350-4. [ Links ]
23. Tabor H, Mehler AH. Histidase and urocanase. Methods in Enzymology 1955; 2: 228-33. [ Links ]
24. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 1977; 74: 5463-7. [ Links ]
25. Okamura H, Nishida T, Nakagawa H. L-histidine ammonia-lyase in rat liver. I. Purification and general characteristics. J Biochem 1974; 75: 139-52. [ Links ]
26. Brand LM, Harper AE. Histidine ammonia-lyase from rat liver. Purification, properties, and inhibition by substrate analogues. Biochemistry 1976; 15: 1814-21. [ Links ]
27. White PJ, Kendrick KE. Inactivation of histidine ammonia-lyase from Streptomyces griseus by dicarbonyl reagents. Biochim Biophys Acta 1993; 1163: 273-9. [ Links ]
28. Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr 1996; 16: 179-203. [ Links ]
29. Jungermann K, Kietzmann T. Role of oxygen in the zonation of carbohydrate metabolism and gene expression in liver. Kidney Int 1997; 51: 402-12. [ Links ]
30. Miyanaka K, Gotoh T, Nagasaki A, Takeya M, Ozaki M, Iwase K, Takiguchi M, Iyama KI, Tomita K, Mori M. Immunohistochemical localization of arginase II and other enzymes of arginine metabolism in rat kidney and liver. Histochem J 1998; 30: 741-51. [ Links ]