Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Botanical Sciences
versión On-line ISSN 2007-4476versión impresa ISSN 2007-4298
Bot. sci vol.92 no.1 México mar. 2014
Revisión
A review of the effect of relaxed clock method, long branches, genes, and calibrations in the estimation of angiosperm age
Una revisión del efecto de los métodos de reloj molecular relajado, ramas largas, genes y calibraciones en la estimación de la edad de las angiospermas
Susana Magallón
Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City, Mexico. Email: s.magaNon@ib.unam.mx
Received: January 29th, 2013
Accepted: September 5th, 2013
Abstract
The timing of angiosperm origin and diversification is crucial to understand the evolution of terrestrial ecosystems as we know them today. The fossil record and relaxed molecular clocks provide substantially different estimates of the time in which angiosperms started to diversify into lineages that originated living species. The roles of different methodological and systematic factors on ages estimated with relaxed clocks across land plants, with a focus on angiosperms, are here reviewed. Different relaxed clocks and molecular data provided similar age estimates for nodes across land plants. Breaking the long branch subtending angiosperms typically did not result in substantially younger age estimates for angiosperms. Including or excluding fossil-derived calibrations on internal nodes had a pronounced effect on ages across the tree, and among examined factors, calibrations were the most influential in resulting ages. However, interactions between calibrations and genes were detected as different genes converged on similar ages when the same set of calibrations was applied. Estimated angiosperm ages were much older than angiosperm earliest fossils. Although a missing fossil history cannot be ruled out, this explanation alone is insufficient, considering the gradually increasing abundance, diversity and geographical distribution of angiosperms in the fossil record; the ordered progression of their morphological and functional diversification; and the agreement in the sequence of appearance of lineages in the stratigraphic record and in molecular phylogenies. It is hypothesized that model misspecification may play a role in the observed molecular clock/fossil discrepancy.
Key words: calibrations, fossil record, land plants, lineage effects, model misspecification.
Resumen
La edad del origen y diversificación de las angiospermas son cruciales para entender la evolución de los ecosistemas terrestres modernos. El registro fósil y los relojes moleculares relajados calculan edades substancialmente diferentes para el inicio de la diversificación de las angiospermas. Se revisa el efecto de diferentes factores metodológicos y sistemáticos en los estimados de relojes relajados para las plantas terrestres, con énfasis en las angiospermas. Diferentes métodos de reloj relajado y datos moleculares estimaron edades similares en todo el árbol filogenético. Romper la rama larga que subyace a las angiospermas no resultó en edades substancialmente más jóvenes. La inclusión o exclusión de calibraciones derivadas del registro fósil tuvo un efecto pronunciado en las edades estimadas, además de ser el factor más relevante entre los que se evaluaron. Sin embargo, existen interacciones entre las calibraciones y los genes, pues diferentes genes convergieron en edades similares cuando las mismas calibraciones fueron aplicadas. Las edades estimadas para las angiospermas son mucho más viejas que sus primeros fósiles. Aunque la posibilidad de un registro fósil incompleto no puede excluirse, ésta sola explicación es insuficiente, dado el aumento gradual en la abundancia y distribución geográfica de las angiospermas en el registro fósil; la progresión ordenada de su diversificación morfológica y funcional; y la concordancia en la secuencia de aparición de linajes en la secuencia estratigráfica y en las filogenias moleculares. Se sugiere que la especificación errónea del modelo evolutivo posiblemente juega un papel en la discrepancia entre los relojes relajados y el registro fósil.
Palabras clave: calibración, efecto del linaje, modelos mal especificados, plantas terrestres, registro fósil.
Flowering plants are an extraordinary evolutionary phenomenon. With over 280,000 described species (Stevens, 2012) and an estimated total of approximately 400,000 (Govaerts, 2001), flowering plants (angiosperms, Angiospermae, Cantino et al., 2007) are by far the most species-rich group of plants. Their richness is even more impressive compared to that of other seed plants, and considering the time of their origin. Whereas extant gymnosperm lineages originated from the Late Carboniferous to the Early Triassic, between 320 and 245 million years (Ma) ago, and collectively include fewer than 1,000 species in the present-day, angiosperms are known in the fossil record only since approximately 135 to 140 Ma ago, and their species richness surpasses that of gymnosperms, and of any other group of plants, by orders of magnitude. For example, several angiosperm genera include more species than all living gymnosperms together. It is clear that, whatever the combination of speciation and extinction rates that led to their present-day species richness, angiosperms, or more likely, particular lineages within the group (Magallon and Sanderson, 2001) are characterized by high rates of phylogenetic diversification.
In addition to their extraordinary species richness, angiosperms have evolved an astounding diversity of vegetative and reproductive innovations. For example, angiosperms encompass completely new forms of herbaceous vegetative constructions, as well as trees with complex branching systems and dense canopies. Reproductively, angiosperms underwent complex morphological and functional innovations, resulting in flowers, which are highly integrated complex structures that function effectively towards selective reproduction, including a highly modified and reduced life cycle.
Angiosperms are the dominant group of plants in modern terrestrial ecosystems. They are the primary producers in food webs, and provide the main structural support in most terrestrial biomes, involving direct physical interactions with all other forms of terrestrial life. The evolution of angiosperms, and specifically the biomes that they formed, were very likely important triggers to the evolution and diversification of numerous biological lineages, including for example, groups of ferns, fungi, insects and arthropods.
Accurate information about the timing of angiosperm origin and diversification is critical to document when angiosperm unique morphological innovations evolved; how long it took for extant lineages to arise, and for modern terrestrial biomes to assemble. To address these questions, it is important to distinguish between the time when the angiosperm lineage diverged from its sister group (i.e., the stem group age), and the time in which extant lineages originated (i.e., the crown group age). In the rest of this review, "angiosperm age" will refer to the angiosperm crown age, unless otherwise indicated. Two sources of information about angiosperm age are available: the fossil record, and molecular clocks.
The fossil record indicates the time by which a lineage had achieved morphological distinctiveness and sufficient abundance to enter a preservation process that would lead to fossilization and subsequent recognition by biologists. Therefore, inherently, the fossil record does not indicate the time of phylogenetic divergence, but a younger age (Figure 1). The amount of time between phylogenetic divergence and fossil preservation is unknown and varies depending on many factors, including a lineage's rate of morphological evolution, life history and ecological preferences. For example, for organisms with high rates of morphological evolution, hard body parts, and that inhabit environments that facilitate preservation, the difference between phylogenetic divergence and fossil preservation is expected to be short. The fossil record of angiosperms as a whole provides an overview of the evolutionary diversification of the group in terms of increasing geographical distribution, local abundance, morphological and functional vegetative and reproductive diversity, taxonomic diversity, and ecological occupation and eventually, dominance (see below). However, the fossil record of particular angiosperm lineages (e.g., families) is incomplete and is mostly characterized by a single or very few occurrences. The oldest unequivocal ocurrence of angiosperms in the fossil record is documented by pollen grains from latest Valanginian and Hauterivian (Early Cretaceous) sediments (see below), corresponding approximately to 140 to 136 Ma, from localities in Israel (Brenner, 1996), Italy (Trevisan, 1988), and southern England (Hughes and McDougall, 1987; Hughes et al., 1991). Whereas these fossil pollen grains strictly provide a minimum age for the angiosperms, the dense representation of readily recognizable angiosperm fossils in sediments from the Barremian onwards suggests that the angiosperm crown group is unlikely to be much older than the oldest Early Cretaceous angiosperms fossils (Pirie and Doyle, 2012; see Discussion).
Molecular clocks are methods that estimate the time of phylogenetic splitting on the basis of genetic distances in phylogenetic trees, and either rely on the assumption that the rate of molecular substitution is approximately constant, and hence, that the genetic distance between two organisms linearly reflects the time elapsed since their divergence from their most recent common ancestor (e.g., strict molecular clocks); or recognize that different lineages usually exhibit different molecular rates, and attempt to accommodate these different rates through different assumptions of how rates vary among lineages (i.e., relaxed molecular clocks).
Several molecular estimates of angiosperm age are available, derived both from strict and relaxed clocks, and most provide ages that are substantially older than the oldest angiosperm fossils (Table 1). Some of the earliest strict clock studies (e.g., Ramshaw, 1972 Martin et al., 1989, 1993; Brandl et al., 1992) estimated ages of 300 Ma or older for angiosperms, or within angiosperms. Relaxed clock studies have also provided older-than-fossil estimates, for example, the most recent studies (Bell et al., 2010; Magallón, 2010; Smith et al., 2010; Clarke et al., 2011) estimate the crown age of angiosperms between 147 Ma (Bell et al., 2010) and 275 Ma (Magallón, 2010). These studies include differential taxonomic sampling and density, are based on different types of data and implement different calibrations and constraints, hence, it not possible to jointly examine the effect of individual parameters on angiosperm age estimates. However, some studies have evaluated the effect of calibration and constraints (Soltis et al., 2002; Moore et al., 2007; Bell et al., 2010; Smith et al., 2010; Clarke et al., 2011; Magallón et al., 2013) the use of different codon positions (Sanderson and Doyle, 2001; Magallón and Sanderson, 2005; Magallón et al., 2013); of different methods to estimate branch lengths (Soltis et al., 2002; Magallón, 2010), and of different relaxed clock method (Magallón, 2010; Magallón et al., 2013).
A possible explanation for the substantially older ages indicated by molecular clocks with respect to the fossil record of angiosperms, and of other biological groups in which an equivalent conflict has been detected, is that the fossil record is incomplete. in the case of angiosperms, an examination of the similarities and differences between the molecular and phylogenetic estimates, and the pattern of the angiosperm fossil record, suggest that an incomplete fossil record alone is an insufficient explanation for the degree of this discrepancy.
Actually, the angiosperm fossil record and molecular-based estimates of angiosperm evolution are congruent in two very important points: both indicate that the onset of angiosperm diversification was characterized by a rapid phylogenetic radiation; and both recognize the same general sequence of appearance of major angiosperm lineages. However, they differ in the absolute time when the angiosperms started to diversify to give rise to living lineages.
Three observations strongly suggest that the angiosperm early fossil record is not severely incomplete. First, the mode of appearance of angiosperms in the fossil record is congruent with the diversification of a biological lineage: The oldest angiosperm pollen grains are approximately spherical, with a single aperture which may be weak or absent, and a perforate to reticulate tectum. These pollen grains are recognized as angiosperms because they have a collumelar infratectal structure. Not all angiosperms have a collumelar infratectum (some lineages have a granular infratectum or different special conditions), but among the living plants, a columellar infratectum is present only in angiosperms (Doyle, 2005, 2009, 2012). The oldest pollen grains with a columellar infratectum are known from a latest Valanginian locality in Israel (Brenner, 1996) and in northern Italy (Trevisan, 1988); and from Hauterivian sediments in southern England (Hughes and McDougall, 1987; Hughes et al., 1991). Angiosperm pollen grains are extremely scarce in each of these localities, being known from a single or very few grains in samples with an otherwise rich palynomorph composition. Palynofloras from slightly younger sediments, specifically of Barremian age, from several places around the world (e.g., Portugal, China, eastern North America) contain angiosperm pollen grains in greater abundance and morphological diversity than the Valanginian and Hauterivian localities. A pattern of increasing global geographical distribution, local abundance, and morphological diversity of angiosperm pollen grains continues through the Early and Late Cretaceous, until angiosperm pollen becomes a dominant component of global palynofloras. The increasing geographical distribution and local abundance of angiosperm pollen has a clear latitudinal component, with an initial expansion in paleoequatorial regions, trailed by occupation of higher paleolatitudes (e.g., Lidgard and Crane, 1988; Crane and Lidgard, 1989; Lupia et al., 1999).
Second, the sequence of morphological evolution observed in fossil pollen grains, leaves, and flowers during the Early and early Late Cretaceous (ca. 135-90 Ma) documents an increasing morphological and functional diversity that is congruent with the expansion of a biological lineage: The morphological succession of pollen grains begins with monosulcate (and inaperturate) grains in the Valanginian and Hauterivian; followed by monosulcates and tricolpates in the Aptian and early Albian; followed by tricolporates in the middle Albian; triangular tricolporates in the late Albian; and complex triporates in the Cenomanian (Doyle, 2012). The leaf morphological succession begins with simple forms with a pinnate but irregular venation pattern, a few palmately-veined, and ternately dissected leaves in the Aptian to earliest Albian; followed by more complex types such as palmately-veined cordate and peltate leaves, pinnately-veined with toothed margin, palmately-lobed, and pinnately-dissected and compound leaves in the middle to late Aptian (Doyle, 2012). Floral evolution also shows an unequivocal morphological and functional progression in reproductive, protective and attractive functions, from very simple types apparently consisting only of stamens and carpels (reproductive function) in the Early Cretaceous; followed by flowers with tepals (protective function) at the end of the Early Cretaceous; followed by flowers with a perianth differentiated into sepals and petals (protective and attractive functions), as well as stamens that have become modified into nectaries (attractive function) by the earliest Late Cretaceous. Younger sediments contain an astounding diversity of floral morphological and functional types (e.g., Friis et al., 2006). The morphological progression of pollen, leaf and flower types observed in the stratigraphic sequence would be hard to explain if angiosperms had evolved a long time before their first fossil appearance.
Third, there is a strong agreement in the general sequence of appearance of major angiosperm lineages in the stratigraphic record and the sequence of branching in molecular phylogenies: The oldest fossil floras that contain reproductive remains with sufficiently detailed preservation to allow comparisons with living lineages occur in localities from Portugal (e.g., Friis et al., 2004), Gabon (Doyle et al., 1990a, b), eastern North America (e.g., Crane et al., 1993; von Balthazar et al., 2007) and Brazil (e.g., Mohr and Bernardes-de-Oliveira, 2004), dated from the late Barremian to the middle Albian (between ca. 125 and 106 Ma). These localities contain exquisitely three-dimensionally preserved flowers, fruits, and seeds, or well-preserved reproductive and vegetative organs in more conventional types of fossilization. These Early Cretaceous paleofloras contain numerous plants that cannot be assigned to extant lineages, and that most likely belong to extinct phylogenetic branches. However, these early floras also contain a few fossils can be reliably assigned, both on the basis of morphological and structural detailed similarity and explicit phylogenetic results (e.g., Friis et al., 2009; Doyle and Endress, 2010), to particular extant lineages. It is extraordinarily significant that the oldest identifiable angiosperm fossils correspond to the earliest diverging branches in molecular phylogenies (Doyle, 2012), specifically, Nymphaeales (Monetianthus mirus; Friis et al., 2001, 2009); Austrobaileyales (Anacostia spp.; Friis et al., 1997a); Chloranthales (the Asteropollis plant; Friis et al., 1994a, 1997b, 1999); the earliest diverging branches within Magnoliidae, namely Calycanthaceae and Lauraceae within Laurales (Virginianthus calycanthoides; Friis et al., 1994b and Potomacanthus lobatus, Crane et al., 1994; von Balthazar et al., 2007, respectively), Magno-liaceae within Magnoliales (Endressinia brasiliana; Mohr and Bernardes-de-Oliveira, 2004), and Winteraceae within Canellales (Walkeripollis; Doyle et al., 1990a, b); Mono-cotyledoneae, namely Araceae within Alismatales (Mayoa portugallica; Friis et al., 2004); and Eudicotlyledoneae, namely Ranunculales (Teixeiraea lusitanica; von Balthazar, 2005) and Nelumbonaceae and Platanaceae within Proteales (the Nelumbites plant; upchurch et al., 1994, and the Sapindopsis plant; Crane et al., 1993, respectively). Each of these three lines of evidence is substantial in itself, and together, they constitute a formidable objection to the possibility of an extensive missing history in the early fossil record of angiosperms.
In this review, some factors that might affect relaxed molecular clock estimation of angiosperm age are examined. I summarize previous work evaluating the effect of relaxed molecular clock methods; of long phylogenetic branches; of molecular data with different substitution rates; and of fossil-derived temporal calibrations. I conclude by hypothesizing possible causes that might explain the age discrepancy between fossils and molecular clocks.
Material and methods
The materials and methods implemented in each study differ in particular details, but the following generalized description applies to all cases.
Taxonomic sample. Phylogenetic and dating analyses were based on a sample of all the living lineages of vascular plants (Tracheophyta) or land plants (Embryophyta). Angiosperms are represented by placeholders of all their evolutionary lineages, namely Amborellales, Nyphaeales, Austrobaileyales and Mesangiospermae, including representatives of its five lineages: Chloranthales, Magnoliidae, Monocotyledoneae, Ceratophyllales and Eudicotyledoneae. An attempt was made to include species with different growth forms and life cycles, to account for potential biases associated with differential molecular rates. Outside angiosperms, cycads (Cycadophyta), Ginkgo, conifers (Coniferae) and gnetales (Gnetophyta) were included, for a full representation of living seed plants (Spermatophyta). Outside seed plants, the five living lineages of ferns (Monilophyta) were included, to represent euphyllophytes (Euphyllophyta). To include a full sample of vascular plants (Tracheophyta), Lycopodiophyta were also included. In the final analyses (Magallón et al., 2013), the three living non-vascular lineages, namely liverworts (Marchatiophyta), mosses (Bryophyta), and hornworts (Anthocerotophyta) were added. The deepest split within each major land plant clades is represented, allowing the estimation of their stem group and crown group age. When only vascular plants were included, a non-vascular plant was selected as outgroup, and when all land plants were included, the outgroup was a member of streptophyte algae, which have been identified as most closely related to the embryophyte lineage.
Data and model selection. The molecular data are four highly-conserved plastid protein-coding genes selected because they offer the possibility to accurately capture relationships at deep phylogenetic levels among land plants. These four genes are atpB (Hoot et al., 1995),psaA,psbB (Graham and Olmstead, 2000; Sanderson et al., 2000), and rbcL (Chase et al., 1993). Sequences were obtained following conventional PCR and sequencing protocols described in Magallón and Sanderson (2002, 2005), or downloaded from GenBank. Subsequently, to evaluate the effect of data with different rates of molecular substitution in dating analyses, matK, with a higher rate of substitution, was included (Magallón et al., 2013). The sequences of matK were obtained with protocols described by Wicke and Quandt (2009) and Worberg et al. (2007), or obtained from GenBank. Each of the four highly-conserved genes was manually aligned, as they almost entirely lack indels. Alignment of matK involved translating nucleotide into amino acid sequences, due to the prevalence of indels.
The best fitting nucleotide substitution model for the three codon positions of each of the highly-conserved genes, and for matK as a whole, was identified with ModelTest (Posada and Buckley, 2004). Following an examination of ModelTest-estimated parameter values according to best-fitting models, the data was partitioned into first plus second codon positions of the highly-conserved genes, third codon positions for the highly-conserved genes, and matK (Magallón et al., 2013).
Phylogenetic analyses. To evaluate phylogenetic congruence among genes, parsimony analyses for each of the highly-conserved genes (Magallón and Sanderson, 2005), and matK (Magallón et al., 2013), were conducted with PAUP* 4.0b10 (Swofford, 2002). Previous studies (e.g., Chaw et al., 2000; Sanderson et al., 2000; Sanderson and Doyle, 2001; Magallón and Sanderson, 2002; Rydin et al., 2002) have detected systematic biases when all codon positions are included in parsimony phylogenetic estimation among seed plants, in a likely case of long branch attraction. This problem is avoided by excluding third codon positions or using amino acid sequences in parsimony analyses. Phylogenetic relationships derived from first plus second positions of different genes were mostly congruent with each other and with independent estimates of phylogenetic relationships across land plants (e.g., Soltis et al., 2011). Because incongruent relationships derived from different genes are weakly supported by bootstrap values, the four highly-conserved genes and the five genes were concatenated, forming a highly-conserved and an all-gene data set, respectively, both of which were used in parametric phylo-genetic estimation.
Phylogenetic relationships were estimated with Bayesian inference using MrBayes (Huelsenbeck and Ronquist, 2001) and with maximum likelihood (ML) using RAxML (Stamatakis, 2006). Analyses were conducted with the best-fitting model to the data partitions described above (as much as each software permits), allowing unlinked substitution parameter estimation. Details of each analysis are described in Magallón and Sanderson (2005), Magallón (2010) and Magallón et al. (2013). Phylogenetic relationships estimated with Bayesian inference for the highly-conserved data set, and with ML for the highly-conserved, matK, and the all-gene data sets were also highly congruent with each other, and with independent estimates of relationships across vascular plants and land plants. Most clades are supported by high posterior probabilities and bootstrap values, respectively.
Relaxed molecular clocks. Relaxed clocks allow estimation of divergence times and absolute molecular substitution rates using molecular sequence data and phylogenetic information, accounting for different molecular substitution rates among branches in the tree. These methods can incorporate independent information, either of absolute time or substitution rate, to calibrate phylogenetic trees. Currently available methods differ in their statistical basis, in how they account for different molecular substitution rates among different branches in the phylogenetic tree, and in how they implement independent calibrations.
Except for the very first proposed relaxed clock (non-parametric rate smoothing; Sanderson, 1997), all available relaxed clocks estimate substitution rates among branches statistically, either with ML (penalized likelihood: PL; Sanderson, 2002), or with Bayesian inference (e.g., Multidivtime, MCMCTREE, TimeTree, BEAST).
Relaxed clocks require a model of how rates change among branches. Among-lineage rate heterogeneity is commonly applied on the basis of temporal autocorrelation (Gillespie, 1991), which assumes that substitution rates (or the factors that determine substitution rates) are transmitted from ancestors to descendants, and therefore are expected to be similar among closely related lineages. Autocorrelation is based on population genetics, and is a reasonable assumption for rate change among closely related species. However, it may be insufficient to explain rate change among branches that are distantly related, that diverged in the distant past, or that are separated by numerous extinct branches. Drummond et al. (2006) implemented uncorrelated among-lineage rate heterogeneity, in which substitution rates are obtained from a single statistical distribution (e.g., exponential, lognormal).
Independent calibrations, either an absolute time or an absolute substitution rate, are necessary if molecular clocks are expected to provide estimates of divergence times and substitution rates in terms of absolute time units (e.g., million years). Temporal calibrations are more common, and among these, the fossil record is the most usual source. Depending on the method used, temporal calibrations can be implemented as fixed ages or as minimum or maximum age constraints with hard or soft bounds (Yang and Rannala, 2006; Rannala and Yang, 2007).
Ages of clades were estimated with three different relaxed clocks (Magallón and Sanderson, 2005; Magallón, 2010; Magallón et al., 2013). Penalized likelihood (Sanderson, 2002) is a semiparametric method that estimates absolute rates with ML, and implements an autocorrelation-based numerical penalty to avoid rates among close branches to vary drastically. It estimates the optimal degree of among-lineage rate heterogeneity through data-driven cross validations that, either predict length of removed terminal branches (i.e., branch-pruning cross validation) or on avoiding violations to temporal constraints (i.e., fossil-based cross validation; Sanderson, 2004). Temporal calibrations can be implemented as fixed ages or as hard-bound minimum or maximum age constraints.
Multidivtime (MD; Thorne et al., 1998; Kishino et al., 2001; Thorne and Kishino, 2002) is a Bayesian method that estimates divergence times, substitution rates and the value of an autocorrelation hyperparameter that models the amount of rate heterogeneity among branches. These (and other) parameters are estimated as posterior probabilities on the basis of priors, a likelihood function, and a Markov chain Monte Carlo (MCMC). This method takes advantage of multigene data sets by considering that, whereas different genes may have uncorrelated patterns of rate change, they are expected to share divergence times (Thorne and Kishino, 2002). Temporal calibrations can be implemented on selected nodes as hard-bound minimum or maximum age constraints, and additionally, a maximum age needs to be assigned to the root of the phylogenetic tree.
The program BEAST (Drummond et al., 2006) offers a variety of methods in a Bayesian framework to estimate divergence times and substitution rates, including implementations with uncorrelated rate change among branches (see above). Phylogenetic relationships, divergence times and absolute substitution rates are estimated simultaneously as posterior probabilities via MCMCs. The method allows great flexibility to implement temporal calibrations, which are introduced as priors drawn from statistical distributions, with hard or soft bounds at either end. We implemented the uncorrelated lognormal method (UCLN) to estimate divergence times and absolute rates.
Calibrations and constraints. Fossil-derived calibrations were implemented in almost all cases. The relationship of fossils with respect to living taxa was derived, either from direct morphology-based phylogenetic analyses (e.g., Kenrick and Crane, 1997; Hilton and Bateman, 2006; Doyle and Endress, 2010) or on the presence of characters in the fossil that constitute unequivocal synapomorphies of a particular clade. The root node was calibrated either with the age of the oldest fossil remains of vascular plants or with a time range derived from an independent dating analysis (Hackett et al., 2007). Table 2 lists the calibrated nodes and the fossils used for calibration.
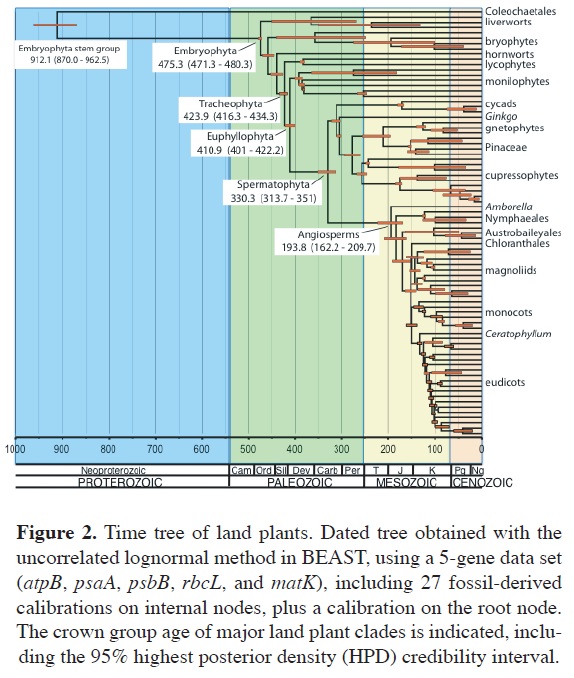
A timeline of land plant evolution. The relaxed clock analyses provided a framework of the time of lineage origin and diversification across land plants. Figure 2 shows the dated tree for land plants estimated with the UCLN method in BEAST, using the all-gene data set (Magallon et al., 2013). The divergence between streptophyte algae and land plants is dated at 912 (962.5-870 95% Highest Posterior Density [HPD]) Ma, corresponding to the Neoproterozoic, and the onset of land plant diversification is estimated at 475.3 (480.3-471.3 95% HPD) Ma, in the Lower Ordovician. Thus, according to these estimates, land plants persisted as an aquatic stem lineage for about 450 Ma before diversifying on land. Vascular plants began their crown diversification at 424 (434.3-416.3 95% HPD) Ma, corresponding to the Middle Silurian. The crown age of euphyllophytes is estimated as 411 (422.2-401 95% HPD) Ma, corresponding to the Lower Devonian, followed relatively soon by fern crown diversification at 392.3 (400.6-384.8 95% HPD) Ma, in the Middle Devonian. Seed plant crown diversification began later, at 330.3 (351-313.7 95% HPD) Ma, in the Lower Carboniferous (Mississippian). Angiosperm crown diversification is estimated at 193.8 (209.7-162.2 95% HPD) Ma, in the Lower Jurassic (Figure 3). Ages estimated for all major land plant clades are consistent with the implemented fossil-based calibrations, except for angiosperms, which are estimated as being ca. 57 Ma older than their earliest fossils (Magallon et al., 2013).
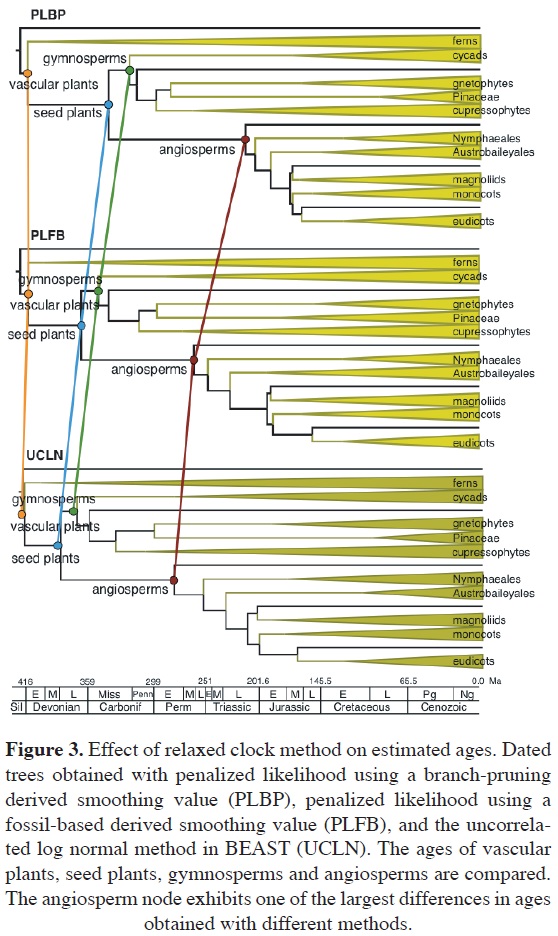
Effect of relaxed clock method. Among the available studies of angiosperm age, few have been designed to examine the effect of different molecular clock methods (Magallon, 2010; Magallon et al., 2013; Table 1). In the study of Magallon (2010), the effect of relaxed clocks that differ in their statistical foundation, in how they implement among-line-age rate heterogeneity, and in the flexibility to implement fossil-derived calibrations was evaluated on the ages of nodes across vascular plants. Penalized likelihood (Sanderson, 2002) and Multidivtime (e.g., Thorne and Kishino, 2002) implement autocorrelated rate change among branches, and hard-bounded minimum or maximum age calibrations. The uncorrelated lognormal (UCLN) method in BEAST (Drum-mond et al., 2006) implements uncorrelated among-lineage rate change and temporal calibrations as statistically-distributed priors (lognormal in the reviewed analyses). Penalized likelihood uses ML and a numerical penalty to estimate dates and absolute rates, whereas Multidivtime and UCLN estimate ages and rates (and other parameters) as posterior probabilities, given priors and a likelihood function, via MCMCs.
Ages across the tree estimated with the PL, MD and UCLN relaxed clocks, including different degrees of among-lineage rate heterogeneity derived from branch-pruning and fossil-based cross validations in PL (PLBP and PLFB, respectively), were in general similar to each other (Figure 3). However, there were relatively pronounced differences in the age of angiosperms estimated by different clocks, namely, 215.6 (202.29-226.77 95% HPD) Ma with PLBP; 260.33 (232.37-284.33 95% HPD) Ma with PLFB; 253.22 (231.45275.86 95% HPD) Ma with MD; and 275 (200-332.37 95% HPD) Ma with UCLN (Magallon, 2010). All estimated angiosperm ages are much older than the earliest fossils of the group. Ages estimated with different relaxed clocks for other nodes across the tree are highly correlated, with the most similar ages estimated with PLFB and MD (correlation r = 0.99 and slope m = 1.0).
The finding that examined relaxed clocks estimated very similar ages across vascular plants was encouraging because the methods are based on different assumptions about among-lineage molecular rate heterogeneity, and on how temporal constraints are implemented. The observation that the most pronounced age differences between different methods occur in nodes that are distant from calibrations, and vice versa, lead to the suspicion that calibrations might be forcing different relaxed clocks into estimating similar ages. To evaluate this possibility, ages estimated with PLBP and UNCL, excluding temporal constraints, were compared (Magallon et al., 2013). Excluding fossil constraints had a substantial effect on the age of angiosperms estimated by PLBP and UNCL, being similar when calibrations were included (i.e., 201.16 and 193.76 Ma respectively), but much different when calibrations were excluded (i.e., 161.64 and 201.02 Ma respectively; Magallon et al., 2013). However, for most other nodes across the tree, the difference between ages estimated with different clocks was small. Ages estimated with the two methods are highly correlated (r = 0.99), but UCLN ages are somewhat older (m = 0.71; Magallon et al., 2013). Thus, in the studies described above the method of relaxed clock had an insubstantial effect on age estimates, regardless of whether constraints were included or not. However, in other studies (e.g., Perez-Lozada et al., 2004) relaxed clock methods had a substantial influence on estimated ages.
Effect of long branches. The effect of long branches in molecular dating has been scantly considered. The effect of long branches in land plant dating was discussed by Sanderson and Doyle (2001), and experimentally evaluated in angiosperm dating by Magallon (2010). A long phylogenetic branch separates the crown node of seed plants from the crown node of angiosperms. This long-branch has been considered a possible cause for the old age estimated by relaxed clocks. Long branches are a well-known source of phylogenetic error (e.g., Felsenstein, 1978; Hendy and Penny, 1989); specifically, with parsimony, homoplasy is mistakenly interpreted as homology; and in parametric phylogeny estimation they may be problematic when models are misspecified. However, very little is known about the effect of long branches in relaxed clock analyses. At least in theory, accurately modeling the rate of substitution on a long branch is challenging due to the greater number of plausible combinations of elapsed time and absolute rate. Nevertheless, there are no reasons to expect that long branches will systematically result in older age estimates for the node they subtend.
If the long branch is responsible for the old relaxed clock angiosperm age, we would expect that ages estimated after the long branch was subdivided into shorter segments (i.e., broken) would be younger. A long branch can be broken by expanding the taxonomic sample to include taxa that diverge from the long-branch, effectively transforming a single long branch into several small ones. While this approach is simple, it cannot be straightforwardly applied to angiosperms, because all the taxa that diverge from their stem branch are extinct, and molecular sequence data for these lineages is unavailable. To confront this situation, a simulations approach was implemented.
First, it became necessary to identify the extinct seed plant lineages that diverged from the angiosperm stem branch (i.e., the angiosperm stem relatives; Figure 4A). This was accomplished through a parsimony phylogenetic analysis including representatives of the major living and extinct seed plant lineages, and combining morphological data (Doyle, 2006) for all taxa, with the highly-conserved molecular data for living taxa. The strict consensus of 30 most parsimonious trees is almost entirely resolved, and it identifies three angiosperm stem relatives (Figure 4A). The earliest diverging lineage leads to Glossopteris (Glossopteridales; Permian Gondwanan "seed ferns") plus Pentoxylon (Pentoxylales; Jurassic-Cretaceous gymnosperms). The second lineage is Bennettitales (Triassic-Cretaceous gymnosperms vegetatively similar to cycads, but with different reproductive organs). The third lineage, corresponding to the sister group of angiosperms, is Caytonia (Caytoniales; Upper Triassic-Cretaceous "seed ferns"). These fossil seed plant lineages represent a working hypothesis of angiosperm stem relatives (Figure 4A).
The second step involves simulating molecular sequences for angiosperm stem relatives. To achieve this, the mean branch length between the seed plant crown node and all extant terminals, considering the highly-conserved data set on the maximum a posteriori (MAP) Bayesian phylogram, was calculated. The identified angiosperm stem relatives were inserted on the angiosperm stem branch in the MAP phylogram, and each was assigned the calculated mean branch length, but reduced considering the age of their extinction in the fossil record (Magallon, 2010). This approach implicitly assumes that the relative substitution rate of the fossil lineages was average among seed plants, and that this rate did not change through time. Both assumptions are very strong. As a result, a tree including the fossil lineages that are most closely related to angiosperms, with branches in terms of expected relative substitution rates, is obtained (Figure 4B). In this tree, the long branch subtending angiosperms is broken into three smaller branches. Using this phylogram, molecular sequences were simulated with the program SeqGen (Rambaut and Grassly, 1997), implementing the best-fitting substitution model estimated with ModelTest (see General Materials and Methods), and ML-estimated model parameters obtained with PAUP* for the living taxa. This resulted in a data set of simulated sequences for all the terminals in the phylogenetic tree, including the angiosperm fossil relatives (Figure 4B).
The third step was to use the simulated sequences to estimate divergence times across vascular plants with relaxed clocks (Figure 4C; see Relaxed molecular clocks). Ages were estimated using PLBP, PLFB, MD and UCLN, and each was compared with estimates obtained with the same method using the original highly-conserved data and taxonomic sample excluding the fossil lineages.
With three out of four relaxed clock methods, breaking the angiosperm long-branch resulted in a younger angiosperm age. The greatest reduction was obtained with PLFB, in which the mean angiosperm age was 260.33 Ma with the original data set, and at 230.97 Ma when angiosperm stem relatives were included (Figure 5). The smallest difference was obtained with UCLN, where the mean angiosperm age with and without the long-branch was 275 and 274.43 Ma, respectively (Fig. 5). Only PLBP estimated an older angiosperm age when stem relatives were included (221.47 Ma including fossils, and 215.6 Ma with the intact branch). Breaking the long-branch subtending angiosperms did not result in substantially different ages for other nodes in the tree.
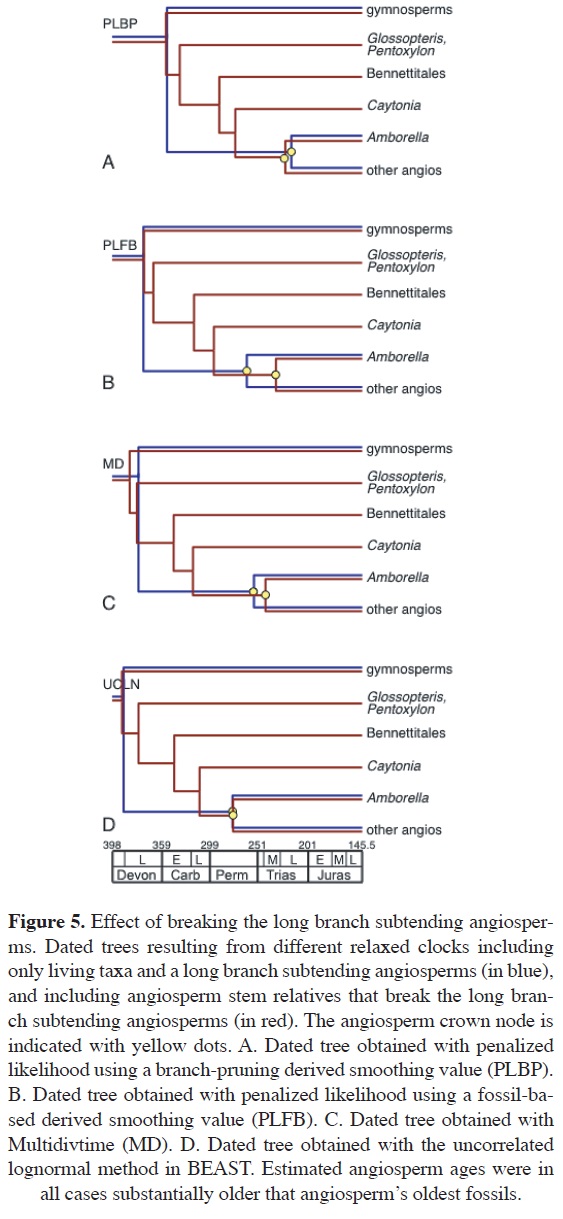
These results indicate that, whereas breaking a long branch can lead to a younger age for the node it subtends, the resulting ages usually do not differ substantially from those obtained with the intact branch. In the particular case of angiosperms, none of the ages obtained after breaking the long-branch was even remotely close to the fossil age. For example, the PLFB method produced the greatest difference in angiosperm age when the long-branch was broken, but nevertheless, this younger age is approximately 90 Ma older than the earliest angiosperm fossils. Although these results did not illuminate a possible cause of the incongruence between angiosperm fossil and relaxed clock age, they are methodologically encouraging, as they suggest that long branches do not have a decisive effect on age estimation. This is good news for dating studies with incomplete taxonomic sampling, or in cases where substantial extinction has taken place.
Gene effects. The relative rate of molecular substitution of nucleotides or amino acids is one of the fundamental sources of information from which divergence times and absolute molecular rates are estimated. Different molecular markers have different relative substitution rates due to differentially conserved functions (e.g., protein-coding, spacers, structural proteins, introns and exons), the genomic compartment they occupy (e.g., in plants, molecular markers from the nuclear genome have higher rates than those from the plastid or mitochondrial genomes), and particular substitution regimes. Although the relative substitution rate of particular genes is likely to be an important factor in reconstructing divergence dates, it would nevertheless be expected that orthologous molecular markers with different relative rates and substitution regimes would provide congruent estimates of ages of clades, because their divergence history derives from the same history of phylogenetic splitting.
Although some previous studies show that different markers estimate different ages (e.g., Sanderson and Doyle, 2001; Soltis et al., 2002), gene effects on relaxed clocks has not been widely evaluated (Table 1), in particular by holding other parameters constant, or involving deep phylogenetic levels. To examine the effect of molecular markers with different substitution regimes and molecular rates on relaxed clock dating, we compared ages derived from two substantially different sets of molecular markers. One is the highly-conserved data set, consisting of atpB, psaA, psbB, and rbcL (see Data and model selection). Each of these genes has a distinct codon structure (Magallon and Sanderson, 2001, 2005), and have been used separately or in different combinations to estimate phylogenetic relationships among land plant lineages that diverged in the distant past. The second data set consists of matK, another protein-coding, plastid gene, but with an overall high rate of molecular substitution, and lacks a marked substitutional differentiation among codon positions (Hilu and Alice, 1999; Hilu et al., 2003). The gene matK has been mostly used to estimate phylogenetic relationships among closely related angiosperm taxa, e.g., within families or genera, but it has also been used to estimate an angiosperm-wide phylogeny (Hilu et al., 2003).
Phylogenetic relationships estimated with the two data sets were equal, except for a few weakly supported nodes, indicating that matK provides phylogenetic signal at deep levels among land plants that is congruent with that provided by highly-conserved plastid genes. The relative substitution rates of matK are ca. 2.5 times higher than those of the highly-conserved genes, but the pattern of branch lengths is preserved in the two phylograms: taxa with long branches in the highly-conserved phylogram also have long branches in the matK phylogram, and vice versa (Magallon et al., 2013). In other words, the pattern of relative substitution rates among lineages is preserved in the two data sets, although the magnitude of the rates is very different.
The two examined data sets provided very similar ages for nodes across the phylogenetic tree, which are highly correlated and proportional (r = 0.99; m = 0.99; Figure 6A). For example, ages estimated with the highly-conserved data set and with matK, respectively for land plants are 474.79 and 474.61 Ma; for vascular plants: 421.78 and 421.65 Ma; for seed plants: 324.35 and 324.69 Ma; and for angiosperms: 208.74 and 194.15 Ma. Credibility intervals associated with these estimates overlapped almost completely (Magallon et al., 2013). Given the high congruence between ages estimated with the two data sets that fit distinct substituion models, we considered the possibility that fossil calibrations were forcing the different data sets to converge on similar ages (as suspected in the case of different relaxed clock methods). To evaluate this possibility, dating analyses excluding the 27 calibrations on internal nodes (but retaining the calibration at the root node), were conducted. It was expected that, by removing calibrations, each data set would estimate substantially different ages across the tree. However, the results did not support this expectation, as unconstrained age estimates across the tree obtained with the highly-conserved and matK data sets were also highly correlated and proportional, although slightly less than when constraints were included (r = 0.98; m = 0.96; Fig. 6B; Magallon et al., 2013).
These results indicate that, in spite of their very different molecular rates and substitution regimes, the two data sets share the same temporal signal. Because the examined sets of genes all belong to the plastid genome, a plausible explanation for the observed proportional branch lengths and very similar divergence times is that different lineages have undergone substitution rate accelerations or decelerations that concertedly affect all the markers in the plastid genome (i.e., lineage effects) in such a way that, within a particular lineage, different markers have low or high rates with respect to other markers in the same lineage, but each marker may consistently exhibit high (or low) rates with respect to the same marker in different lineages.
Note that the similar branch length pattern and age estimates derived from two examined data sets cannot be extrapolated to other molecular data sets, especially if they involve markers from different genomic compartments (i.e., nuclear or mitochondrial). However, if the assumption about the presence of lineage effects is correct, these results would suggest that branch length patterns and age estimates derived from different plastid genes will be similar among land plants.
Calibration effects
Temporal calibrations are critical to estimate ages and absolute substitution rates with relaxed clocks. If correctly interpreted and applied, they provide an absolute temporal framework with which ages and rates in the tree are calibrated. But if incorrectly applied, for example, because of incorrect assignment of a fossil to a clade, or by mistaken stratigraphic placement of a fossil, an erroneous time framework may be propagated across the tree. Previous studies have shown that no amount of molecular data can surmount the temporal framework imposed by calibrations (Yang and Rannala, 2006; Rannala and Yang, 2007). Several dating analyses have examined the effect of calibrations on age estimates across land plants (Table 1; e.g., Soltis et al., 2002; Bell et al., 2010; Smith et al., 2010; Clarke et al., 2011). The analyses described above, in which the effects of relaxed clock method and molecular data were examined, resulted in very similar age estimates for most nodes across the tree when fossil constraints were excluded (Magallon, 2010; Magallon et al., 2013). To explicitly examine the effect of temporal constraints on relaxed clocks, we compared ages across land plants estimated when the set of 27 fossil-derived calibrations was included or excluded, and all other analysis conditions remained equal (Magallon et al., 2013). Calibration effects were evaluated independently using the highly-conserved, matK, and the all-gene data sets. All the major lineages of land plants were included, as well as a strepto-phyte algal outgroup. Divergence dates were estimated with UNCL and PLBP, however because the estimated ages were similar, only the UCLN results will be discussed.
For the three data sets, ages of nodes across the tree obtained including and excluding calibrations were very different (Figure 7). These differences are much larger than those obtained when different relaxed clocks or different genes were used, and more pronounced in nodes subtended by or that subtend long branches. Nevertheless, for any given data set, ages estimated including and excluding calibrations are highly correlated, although unconstrained ages are somewhat older. For example, using the all-gene data set, the correlation and proportionality between constrained and unconstrained ages are r = 0.95 and m = 1.17. Ages including and excluding calibrations for land plants are 475.34 and 701.95 Ma; for vascular plants, 423.95 and 521.54 Ma; for seed plants, 330.33 and 355.52 Ma; and for angiosperms, 193.76 and 201.02 Ma, respectively. The absolute per-node mean difference between unconstrained and constrained ages across the tree is approximately 38 Ma. Furthermore, excluding calibrations substantially increases the size of the credibility interval of estimated ages, with the mean magnitude of unconstrained 95% HPDs being approximately threefold the constrained 95% HPD (Magallon et al., 2013).
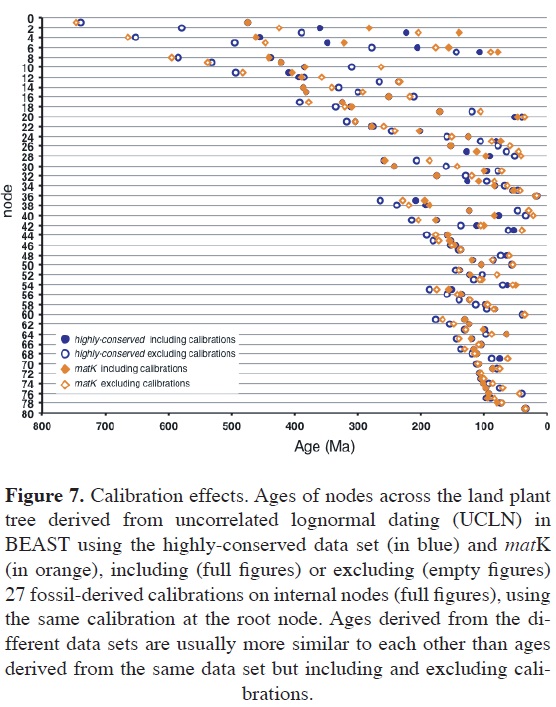
These results clearly support previous theoretical and empirical studies indicating the crucial relevance of calibrations in relaxed clock analyses. Additionally, a comparison among ages estimated with different data sets and including or excluding calibrations revealed unexpected interactions among data sets, which probably result from proportional accelerations or decelerations of the genes in particular lineages: whereas calibrations have the most determinant effect on age estimates, different data sets converged on similar ages if the same set of calibrations was applied, possibly as a consequence of lineage effects.
Discussion
The reviewed studies indicate that different relaxed clocks and different genes did not result in substantially different ages across the tree, and that breaking the long branch subtending angiosperms did not result in substantially different ages for the angiosperm crown node. Including or excluding calibrations to internal nodes, however, had a pronounced effect on ages estimated across the tree. Among the factors examined in this study, calibrations are by far the most influential, an outcome that confirms independent studies documenting the importance of calibrations in relaxed clock age and rate estimation. However, interactions between calibrations and genes were detected: underlying the substantially different ages derived from applying calibrations, different genes converge on similar ages, probably as a consequence of lineage effects.
The studies reviewed above provide an integrative temporal framework of land plant evolution. The ages estimated for major clades across land plants are congruent with their stratigraphic occurrence, except for angiosperms, which are estimated to be much older than their Early Cretaceous fossil appearance. Do these results indicate that the angiosperm crown group is substantially older than the oldest reliable angiosperm fossils? If so, it would imply that the first 50 to 60 Ma of crown angiosperm history did not leave a fossil record. Other studies have also estimated a crown angiosperm age substantially older than its fossil record: such as 147 (141-154) or 183 (167-199) Ma by Bell et al. (2010); 215.6 (202.3-226.8) to 275 (200-332.4) Ma by Magallon (2010); 217 (182-257) Ma, 228 (193-270) Ma by Smith et al. (2010), and 198 (170-231) and 205 (175-240) Ma by Clarke et al. (2011), to cite some recent examples (Table 1).
Another observation that might support a substantially older angiosperm age is that in the UCLN analyses described here (Magallon et al., 2013), the calibration on the angiosperm crown node was implemented as a lognormal prior narrowly distributed around 136 Ma. Nevertheless, the signal in the data overcame this strong prior, to estimate an age that is substantially older and has an extremely low probability given the prior distribution. This result is methodologically interesting, because it unequivocally shows that, if the signal in the data is sufficiently strong, the UNCL method in BEAST it will overcome a strong prior.
These combined observations clearly indicate that the molecular data contain a signal congruent with an angiosperm age much older than the earliest angiosperm fossils. But I would like to argue that a very old angiosperm age is not the only possible interpretation of such molecular signal.
Model misspecification is a well-known source of error in relaxed clock analyses (e.g., Hugall et al., 2007; Lepage et al., 2007; Phillips, 2009; Brandley et al., 2011; Dornburg et al., 2012; Wertheim et al., 2012). Recall that the length of a branch in a phylogenetic tree represents the combination of the absolute rate of molecular substitution and elapsed time. A long branch can result from a high absolute substitution rate, or a long time (or both; Figure 8A), but the absolute rate and time components of a phylogram cannot be separated in a simple way. With relaxed clocks, the absolute rate and time components of a branch are estimated by modeling temporal autocorrelation on branches (e.g., Multidivtime, PL), or by simultaneously modeling both parameters in an uncorrelated context (in BEAST).
I speculate that relaxed clocks systematically miscalculate the absolute rate and absolute time components of the branch subtending the angiosperm crown node, possibly as a consequence of model misspecification. Specifically, I suggest that the length of the branch subtending angiosperms is incorrectly interpreted by relaxed clocks as consisting of a high absolute rate and a moderate elapsed time (Figure 8B), but that in reality, it is derived from a moderate absolute rate and a long elapsed time (Figure 8C). A possible reason for this erroneous optimization may stem from the fact that the long subtending branch is followed by very short branches on the angiosperm backbone.
Two circumstantial observations become relevant. First, although independent relaxed clock estimates of angiosperm age are typically much older that its fossil record (see above), these estimates do not converge on a narrow range, but rather, span between 147 Ma (Bell et al., 2010) and 275 Ma (Magallon, 2010). Therefore, it cannot be considered that relaxed clock estimates have reached a congruent estimation of angiosperm age.
Second, when the age of angiosperms is fixed between 130 and 140 Ma, relaxed clock age estimates within angiosperms are substantially congruent with the fossil record. One case that is particular significant is the eudicot clade (Eudicotyledoneae). Eudicots are morphologically characterized by tricolpate pollen grains (or derived from this condition), and reconstructions of morphological evolution unequivocally indicate that tricolpate pollen evolved on the branch subtending this clade. The oldest fossil tricolpate pollen grains offer an unusually high potential to accurately reflect the time of origin of eudicots, because they represent an unequivocal synapomorphy of the clade, with improved chances of rapidly entering the fossil record, given that pollen (in general) is abundantly produced and is physically and chemically resistant. It is thus, particularly suggestive that when angiosperms as a whole are calibrated to be no older than 140 Ma, relaxed clock ages of stem group and crown group eudicots narrowly bracket the age of the earliest fossil tricolpate pollen grains.
Relaxed clocks are increasingly powerful methods to estimate divergence times and absolute rates. Nevertheless, as any other estimation method, they can provide erroneous results under complex conditions of analysis, or when their assumptions (i.e., models) differ substantially from the evolutionary process underlying the data. Relaxed clocks have been shown to incorrectly estimate divergence dates, for example, as a consequence of overparameterization due to the use of uncorrected gene sequences (Hugall and Lee, 2004); of substitution saturation which leads to rate underestimation and age overestimation (Jansa et al., 2006; Hugall et al., 2007); of the model used to relax the molecular clock (Lepage et al., 2007; and of failure to adequately model DNA substitution, leading to incorrect branch lengths and divergence time estimates (Brandley et al., 2011), which is aggravated with increasing distance from calibration points (Phillips, 2009). Specifically, pronounced substitutional differences among closely related lineages, or between a clade and the rest of the tree, have been shown to systematically provide erroneous estimates of divergence dates (Dornburg et al., 2012; Wertheim et al., 2012). Wertheim et al. (2012) detected the inability of relaxed clocks to capture extensive substitution rate variation (i.e., heterotachy) among closely related lineages. Whereas relaxed clocks were able to detect rate changes in a single clade, the magnitude of the change was substantially underestimated. Through simulations they showed that in cases of extreme rate variation, a single relaxed clock applied to the whole tree cannot adequately capture the magnitude of rate heterogeneity, and leads to age overestimation. Dornburg et al. (2012) found that in the framework of an uncorrelated relaxed clock where different substitution rates are obtained from a single continuous parametric distribution, the presence disparate substitution rates in the tree severely mislead molecular rate estimation. In some cases, especially depending on the number and implementation of calibrations, a rate that is intermediate (but incorrect) between the disparate (but true) rates is estimated, as a consequence of violation of the assumption that the distribution of rates in the tree can be obtained from a single unimodal parametric distribution. The incorrect rates led to poor age estimates. under these conditions, a Bayesian random local clock model (Drummond and Suchard, 2010), which can incorporate disparate rates in different parts of the tree, led to more precise estimates of molecular rate and divergence time (Dornburg et al., 2012). The results from these studies immediately suggest that explicit evaluation of the role of extreme molecular rate variation among angiosperm lineages, and/or of disparate rates between angiosperms and living gymnosperms, should be conducted. Modeling of pronounced rate heterogeneity could be guided by phenotypic attributes correlated with molecular substitution, for example, growth form or life cycle in plants (e.g., Smith and Donoghue, 2008; Baker et al., in press).
Another potentially helpful suggestion is to pay greater attention to the fossil record, specifically, to the different preservation potential of organs and biological lineages; and on the reliability that can be placed on fossilized structures to identify particular clades. This could lead to a more nuanced implementation of fossil-derived calibrations depending qualitative or quantitative assessments of their reliability. For example, Foote et al. (1999) implemented a method based on frequency ratios of fossils, to estimate the probability of a missing fossil history of a given magnitude. More recently, Marshall (2008) proposed a method to determine (with an associated probability) the maximal age of a lineage, based on the overal fossil record of the branches in a phylogenetic tree. We have applied this method to the angiosperms, and found that the maximal age of the angiosperms as a whole is only a few million years older than their oldest fossil record.
Finally, it is important to stress that the extent to which the results found in the studies summarized here regarding the effect of relaxed clocks, branch lengths and genes apply to other data and organisms is unknown. A more extensive examination, with many different conditions of analysis, would be necessary. Some independent studies show different outcomes, specifically, with substantially different estimates resulting from different relaxed clocks or different molecular data. However, all studies have recognized the crucial impact of calibrations on age estimation. All available results indicate that temporal calibrations very possibly represent the single most important factor that determines age estimates in relaxed clock methods. Therefore, greater attention on how best to implement them, and in particular, the differential reliability of fossil-based calibrations, would be useful.
Literature cited
Ash S.R. 1972. Late Triassic plants from the Chinle Formation in northeastern Arizona. Palaeontology 15:598-618. [ Links ]
Baker T., Pennington T., Magallón S., Laurance W., Gloor E., Alexiades M., Alvarez E., Araujo A., Arets E., Aymard G., Oliveira A., Amaral I., Arroyo L., Bonal D., Brienen R., Chave J., Dexter K., DiFiore A., Eler E., Feldpausch T., Ferreira L., López Gonzalez G., van der Heijden G., Higuchi N., Honorio E., Huamantupa I., Killeen T., Laurance S., Toledo M., Lewis,S., Malhi, Y., Marimon B., Marimon Ben H., Monteagudo-Mendoza A., Neill D., Penuela M., Pitman N., Prieto A., Quesada C., Ramirez F., Ramirez-Angulo H., Rudas A., Ruschel A., Salamao R., Andrade A., Silva N., Silveira M., Simon M., Spironelo W., ter Steege H., Terborgh J., Torres A., Vasquez R., Vieira I., Vilanova E., Vos V. and Phillips, O. Fast demographic traits promote high diversification rates of Amazonian trees. Ecology Letters (in press). [ Links ]
Banks H.P. 1980. Floral assemblages in the Siluro-Devonian. In: Dilcher D.L. and Taylor T.N. Eds. Biostratigraphy of fossil plants: Successional and Paleoecological Analyses, pp. 1-24 Dowden, Hutchinson and Ross, Stroudsburg. [ Links ]
Bell C.D., Soltis D.E. and Soltis P.S. 2005. The age of angiosperms: a molecular timescale without a clock. Evolution 59:1245-1258. [ Links ]
Bell C.D., Soltis D.E. and Soltis P.S. 2010. The age and diversification of the angiosperms re-revisited. American Journal of Botany 97:1296-1303. [ Links ]
Brandl R., Mann W. and Sprinzl M. 1992. Estimation of the monocotdicot age through tRNA sequences from the chloroplast. Proceedings of the Royal Society London B 249:13-17. [ Links ]
Brandley M.C., Wang Y., Guo X., Nieto Montes de Oca A., Feria-Ortíz M., Hikida T. and Ota H. 2011. Accommodating heterogeneous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Systematic Biology 60:3-15. [ Links ]
Brenner G.J. 1996. Evidence for the earliest stage of angiosperm pollen evolution: a paleoequatorial section from israel. In: Taylor D.W. and Hickey L.J. Eds. Flowering Plant Origin, Evolution and Phylogeny, pp. 91-115, Chapman and Hall, New York. [ Links ]
Cantino P.D., Doyle J.A., Graham S.W., Judd W.S., Olmstead R.G., Soltis D.E., Soltis P.S. and Donoghue M.J. 2007. Towards a phylogenetic nomenclature of Tracheophyta. Taxon 56:822-846. [ Links ]
Chase M.W., Soltis D.E., Olmstead R.G., Morgan D., Les R.H., Mishler B.D., Duvall M.R., Price R.A., Hills H.G., Qiu Y.L., Kron K.A., Rettig J.H., Conti E., Palmer J.D., Manhart J.R., Sytsma K.J., Michaels H.J., Kress W.J., Karol K.G., Clark W. D., Hedren M., Gaut B.S., Jansen R. K., Kim K.J., Wimpee C.F., Smith J.F., Furnier G.R., Strauss S.H., Xiang Q.Y., Plunkett G.M., Soltis P.S., Swensen S.M., Williams S.E., Gadek PA., Quinn C.J., Eguiarte L.E., Golenberg E., Learn Jr. G.H., Graham S.W., Barrett S.C.H., Dayanandan S. and Albert V.A.. 1993. Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Annals of the Missouri Botanical Garden 80:528-580. [ Links ]
Chaw S.-M., Parkinson C.L., Cheng Y., Vincent T.M. and Palmer J.D. 2000. Seed plant phylogeny inferred from all three plant genomes: monophyly of extant gymnosperms and origin of Gnetales from conifers. Proceedings of the National Academy of Science, USA 97:4086-4091. [ Links ]
Christopher R.A. 1979. Normapolles and triporate pollen assemblages from the Raritan and Magothy Formations (Upper Cretaceous) of New Jersey. Palynology 3:73-121. [ Links ]
Clarke J.T., Warnock R.C.M. and Donoghue PC.J. 2011. Establishing a time-scale for plant evolution. New Phytologist Dryad Digital Repository 192:266-301. [ Links ]
Crane P.R. 1996. The fossil history of the Gnetales. International Journal of Plant Sciences 157(Suppl.):S50-S57. [ Links ]
Crane P.R. and Lidgard S. 1989. Angiosperm diversification and paleolatitudinal gradients in Cretaceous floristic diversity. Science 246:675-678. [ Links ]
Crane PR., Friis E.M. and Pedersen K.R. 1994. Paleobotanical evidence on the early radiation of magnoliid angiosperms. Plant Systematics and Evolution 8(Suppl.):51-72. [ Links ]
Crane P.R., Pedersen K.R., Friis E.M. and Drinnan A.N. 1993. Early Cretaceous (early to middle Albian) platanoid inflorescences associated with Sapindopsis leaves from the Potomac Group of North America. Systematic Botany 18:328-344. [ Links ]
Daghlian C.P 1981. A review of the fossil record of monocotyledons. The Botancial Review 47:517-555. [ Links ]
Delevoryas T. and Hope R.C. 1973. Fertile coniferophyte remains from the Late Triassic Deep River Basin, North Carolina. American Journal of Botany 60:810-818. [ Links ]
Delevoryas T. and Hope R.C. 1987. Further observations on the Late Triassic conifers Compsostrobus neotericus and Voltzia andrewsii. Review of Paleobotany and Palynology 51:59-64. [ Links ]
Dilcher D.L., Sun G., Ji Q. and Li H. 2007. An early infructescence Hyrcantha decussata (comb. nov.) from the Yixian Formation in northeastern China. Proceedings of the National Academy of Sciences, U.S.A. 104:9370-9374. [ Links ]
Dornburg A., Brandley M.C., McGowen M.R. and Near T.J. 2012. Relaxed clock and inferences of heterogeneous patterns of nucleotide substitutions and divergence time estimates across whales and dolphins (Mammalia: Cetacea) Molecular Biology and Evolution 29:721-736. [ Links ]
Doyle J.A. 2000. Paleobotany, relationships, and geographic history of Winteraceae. Annals of the Missouri Botanical Garden 87:303-316. [ Links ]
Doyle J.A. 2005. Early evolution of angiosperm pollen as inferred from molecular and morphological phylogenetic analyses. Grana 44:227-251. [ Links ]
Doyle J.A. 2006. Seed ferns and the origin of angiosperms. Journal of the Torrey Botanical Society 133:169-209. [ Links ]
Doyle J.A. 2009. Evolutionary significance of granular exine structure in the light of phylogenetic analyses. Review of Palae-obotany and Palynology 156:198-210. [ Links ]
Doyle J.A. 2012. Molecular and fossil evidence on the origin of angiosperms. Annual Review of Earth and Planetary Sciences 40:301-326. [ Links ]
Doyle J.A. and Endress PK. 2010. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: Magnoliidae and eudicots. Journal of Systematics and Evolution 48:1-35. [ Links ]
Doyle J.A. and Hotton C.L. 1991. Diversification of early angios-perm pollen in a cladistic context. In: Blackmore S. and Barnes S.H. Eds. Pollen and Spores: Patterns of Diversification, pp. 169-195, Clarendon Press, Oxford. [ Links ]
Doyle J.A., Hotton C.L. and Ward J.V. 1990a. Early Cretaceous tetrads, zonasulculate pollen and Winteraceae. I. Taxonomy, morphology, and ultrastructure. American Journal of Botany 77:1544-1557. [ Links ]
Doyle J.A., Hotton C.L. and Ward J.V. 1990b. Early Cretaceous tetrads, zonasulcate pollen and Winteraceae. II. Cladistic analysis and implications. American Journal of Botany 77:1558-1568. [ Links ]
Doyle J.A., Biens P., Dorenkamp A. and Jardiné S. 1977. Angiosperm pollen from the pre-Albian Cretaceous of Equatorial Africa. Bulletin de Centres de Recherches Exploration-Production Elf-Aquitaine 1:451-473. [ Links ]
Drinnan A.N., Crane P.R., Friis E.M. and Pedersen K.R. 1991. Angiosperm flowers and tricolpate pollen of Buxaceous affinity from the Potomac Group (Mid-Cretaceous) of eastern North America. American Journal of Botany 78:153-176. [ Links ]
Drummond A.J. and Suchard M.A. 2010. Bayesian random local clocks, or one rate to rule them all. BMC Biology 8:114. [ Links ]
Drummond A.J., Ho S.Y.W., Phillips M.J. and Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4:e88. [ Links ]
Fairon-Demaret M. 1974. Noveaux specimens du genre Leclercquia Banks, H. P., Bonamo, P. M., et Grierson, J. D. 1972 du Givetien (?) du Queensland (Australie). Bulletin de l'Institut Royal des Sciences Naturelles de Belgique. Sciences de la Terre 50:1-4. [ Links ]
Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology 27:401-410. [ Links ]
Florin R. 1951. Evolution in cordaitales and conifers. Acta Horti Bergiani 15:285-388. [ Links ]
Foote M., Hunter J.P, Janis C.M. and Sepkoski J.J. Jr. 1999. Evolutionary and preservational constraints on origins of biologic groups: divergence times of eutherian mammals. Nature 283:1310-1314. [ Links ]
Friis E.M., Crane P.R. and Pedersen K.R. 1997a. Anacostia, a new basal angiosperm from the Early Cretaceous of North America and Portugal with trichotomocolpate/monocolpate pollen. Grana 36:225-244. [ Links ]
Friis E.M., Crane PR. and Pedersen K.R. 1997b. Fossil history of magnoliid angiosperms. In: Iwatsuki K. and Raven P.H. Eds. Evolution and Diversification of Land Plants, pp. 121-156, Springer Verlag, Tokyo. [ Links ]
Friis E.M., Pedersen K.R. and Crane P.R. 1994a. Angiosperm floral structures from the Early Cretaceous of Portugal. Plant Systematics and Evolution 8(Suppl.):31-49. [ Links ]
Friis E.M., Pedersen K.R. and Crane PR. 1999. Early angiosperm diversification: the diversity of pollen associated with angiosperm reproductive structures in Early Cretaceous floras from Portugal. Annals of the Missouri Botanical Garden 86:259-296. [ Links ]
Friis E.M., Pedersen K.R. and Crane PR. 2001. Fossil evidence of water lilies (Nymphaeales) in the Early Cretaceous. Nature 410:357-360. [ Links ]
Friis E.M., Pedersen K.R. and Crane P.R. 2004. Araceae from the Early Cretaceous of Portugal: evidence on the emergence of monocotyledons. Proceedings of the National Academy of Sciences, USA 101:16565-16570. [ Links ]
Friis E.M., Pedersen K.R. and Crane P.R. 2006. Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Palaeogeography, Palaeoclimatology and Palaeoecology 232:251-293. [ Links ]
Friis E.M., Eklund H., Pedersen K.R. and Crane PR. 1994b. Virginianthus calycanthoides gen. et sp. nov.- a calycanthaceous flower from the Potomac Group (Early Cretaceous) of eastern North America. International Journal of Plant Sciences 155:772-785. [ Links ]
Friis E.M., Pedersen K.R, von Balthazar M., Grimm G.W. and Crane P.R. 2009. Monetianthus mirus gen. et sp. nov., a Nymphalean flower from the Early Cretaceous of Portugal. International Journal of Plant Sciences 170:1086-1101. [ Links ]
Garrat M.J. and Rickards R.B. 1987. Pridoli (Silurian) graptolites in association with Baragwanathia (Lycophytina). Bulletin of the Geological Society of Denmark 35:135-139. [ Links ]
Gensel P.G. and Andrews H.N. 1984. Plant Life in the Devonian. Praeger, New York. [ Links ]
Gillespie J.H. 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford. [ Links ]
Goremykin V.V., Hansmann S. and Martin W.F. 1997. Evolutionary analysis of 58 proteins encoded in six completely sequenced chloroplast genomes: revised molecular estimates of two seed plant divergence times. Plant Systematics and Evolution 206:337-351. [ Links ]
Govaerts R. 2001. How many species of seed plants are there? Taxon 50:1085-1090. [ Links ]
Gradstein F. and Ogg J. 2004. Geologic time scale 2004 - why, how and where next! Lethaia 37:175-181. [ Links ]
Graham S.W. and Olmstead R.G. 2000. Utility of 17 chloroplast genes for inferring the phylogeny of the basal angiosperms. American Journal of Botany 87:1712-1730. [ Links ]
Hackett J.D., Yoon H.S., Reyes-Prieto A., Rümmele S.E. and Bhattacharya D. 2007. Phylogenetic analysis supports the monophyly of cryptophytes and haptophytes and the association of Rhizaria with Chromalveolates. Molecular Biology and Evolution 24:1702-1713. [ Links ]
Harris T.M. 1961. The fossil cycads. Palaeontology 4:313-323. [ Links ]
Hendy M.D. and Penny D. 1989. A framework for the quantitative study of evolutionary trees. Systematic Zoology 38:297-309. [ Links ]
Hilton J. and Bateman R.M. 2006. Pteridosperms are the backbone of seed-plant phylogeny. Journal of the Torrey Botanical Society 133:119-168. [ Links ]
Hilu K.W. and Alice L.A. 1999. Evolutionary Implications of matK indels in Poaceae. American Journal of Botany 86:1735-1741. [ Links ]
Hilu K.W., Borsch T., Müller K., Soltis D.E., Soltis PS., Savolainen V., Chase M.W., Powell M.P, Alice L.A., Evans R., Sauquet H., Neinhuis C., Slotta T.A.B., Rohwer J.G., C.S. Campbell and Chatrou L.W.. 2003. Angiosperm phylogeny based on matK sequence information. American Journal of Botany 90:1758-1776. [ Links ]
Hueber F.M. 1992. Thoughts on the early lycopsids and zostero-phylls. Annals of the Missouri Botanical Garden 79:474-499. [ Links ]
Huelsenbeck J.P. and Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [ Links ]
Hugall A.F. and Lee M.S. 2004. Molecular claims of Gondwanan age for Australian Agamid Lizards are untenable. Molecular Biology and Evolution 21:2102-2110. [ Links ]
Hugall A.F., Foster R. and Lee M.S.Y. 2007. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Systematic Biology 56:543-563. [ Links ]
Hughes N.F. and McDougall A.B. 1987. Records of angiospermid pollen entry into the English Early Cretaceous succession. Review of Palaeobotany and Palynology 50:255-272. [ Links ]
Hughes N. F. and McDougall A.B. 1990. Barremian-Aptian angiospermid pollen records from southern England. Review of Palaeobotany and Palynology 65:145-151. [ Links ]
Hughes N.F., McDougall A.B. and Chapman J.L. 1991. Exceptional new record of Cretaceous Hauterivian angiospermid pollen from southern England. Journal of Micropalaeontology 10:75-82. [ Links ]
Jansa S.A., Barker F.K. and Heaney L.R. 2006. The pattern and timing of diversification of Philippine endemic rodents: evidence from mitochondrial and nuclear gene sequences. Systematic Biology. 55:73-88. [ Links ]
Kenrick P. and Crane P.R. 1997. The Origin and Early Diversification of Land Plants: A Cladisitc Study. Smithsonian Institution Press, Washington. [ Links ]
Kishino H., Thorne J.L. and Bruno W.J. 2001. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Molecular Biology and Evolution 18:352-361. [ Links ]
Knobloch E. and Mai D.H. 1986. Monographie der Früchte und Samen in der Kreide von Mitteleuropa. Rozpravy Ustfedniho üstavu geologického, Praha 47:1-219. [ Links ]
Krassilov V.A. 1982. Early Cretaceous flora of Mongolia. Palaeontographica Abteilung B: Paläophytologie 181:1-43. [ Links ]
Krassilov V.A. 1986. New floral structure from the Lower Cretaceous of Lake Baikal area. Review of Palaeobotany and Palynology 47:9-16. [ Links ]
Krassilov V.A. and Bugdaeva E.V. 1982. Achene-like fossils from the Lower Cretaceous of the Lake Baikal area. Review of Palaeobotany and Palynology 36:279-295. [ Links ]
Laroche J., Li P. and Bousquet J. 1995. Mitochondrial DNA and monocotdicot divergence time. Molecular Biology and Evoltuion 12:1151-1156. [ Links ]
Lefeld J. 1978. Mongolia. In: Moullade M. and Nairn A.E.M. Eds. The Phanerozoic Geology of the World. II. The Mesozoic A, pp. 55-78, Elsevier Science Publishing, New York. [ Links ]
Leng Q. and Friis E.M. 2003. Sinocarpus decussatus gen. et sp. nov., a new angiosperm with basally syncarpous fruits from the Yixian Formation of Northeast China. Plant Systematics and Evolution 241:77-88. [ Links ]
Leng Q. and Friis E.M. 2006. Angiosperm leaves associated with Sinocarpus infructescences from the Yixian Formation (mid-Early Cretaceous) of NE China. Plant Systematics and Evolution 262:173-187. [ Links ]
LePage B.A. and Basinger J.F. 1995. Evolutionary history of the genus Pseudolarix Gordon (Pinaceae). International Journal of Plant Sciences 156:910-950. [ Links ]
Lepage T., Bryant D., Phillippe H. and Lartillot N. 2007. A general comparison of relaxed molecular clock models. Molecular Biology and Evolution 24:2669-2680. [ Links ]
Lidgard S. and Crane P.R. 1988. Quantitative analysis of the early angiosperm radiation. Nature 331:344-346. [ Links ]
Lupia R., Lidgard S. and Crane PR. 1999. Comparing palynological abundance and diversity: implications for biotic replacement during the Cretaceous angiosperm radiation. Paleobiology 25:305-340. [ Links ]
Magallón S. 2010. Using fossils to break long branches in molecular dating: a comparison of relaxed clocks applied to the origin of angiosperms. Systematic Biology 59:384-399. [ Links ]
Magallón S. and Castillo A. 2009. Angiosperm diversification through time. American Journal of Botany 96:349-365. [ Links ]
Magallón S. and Sanderson M.J. 2001. Absolute diversification rates in angiosperm clades. Evolution 55:1762-1780. [ Links ]
Magallón S. and Sanderson M.J. 2002. Relationships among seed plants inferred from highly conserved genes: sorting conflicting phylogenetic signals among ancient lineages. American Journal of Botany 89:1991-2006. [ Links ]
Magallón S.A. and Sanderson M.J. 2005. Angiosperm divergence times: the effect of genes, codon positions, and time constraints. Evolution 59:1653-1670. [ Links ]
Magallón S., Hilu K.W. and Quandt D. 2013. Land plant evolutionary timeline: gene effects are secondary to fossil constraints in relaxed clock estimation of age and substitution rates. American Journal of Botany 100:556-573. [ Links ]
Marshall C.R. 2008. A simple method for bracketing absolute divergence times on molecular phylogenies using multiple fossil calibration points. American Naturalist 171:726-742. [ Links ]
Martin W., Gierl A. and Saedler H. 1989. Molecular evidence for pre-Cretaceous angiosperm origins. Nature 339:46-48. [ Links ]
Martin W., Lydiate D., Brinkmann H., Forkmann G., Saedler H. and Cerff R. 1993. Molecular phylogenies in angiosperm evolution. Molecular Biology and Evolution 10:140-162. [ Links ]
Miller C.N. Jr. 1971. Evolution of the fern family Osmundaceae based on anatomical studies. Contributions from the Museum of Paleontology, University of Michigan 23:105-169. [ Links ]
Mohr B.A.R. and Bernardes-de-Oliveira M.E.C. 2004. Endressinia brasiliana, a magnolialean angiosperm from the Lower Cretaceous Crato Formation (Brazil). International Journal of Plant Sciences 165:1121-1133. [ Links ]
Moore M.J., Bell C.D., Soltis PS. and Soltis D.E. 2007. Using plastid genome-scale data to resolve engimatic relationships among basal angiosperms. Proceedings of the National Academy of Sciences U.S.A. 104:19363-19368. [ Links ]
Nixon K.C. and Crepet W.L. 1993. Late Cretaceous fossil flowers of ericalean affinity. American Journal of Botany 80:616-623. [ Links ]
Pacltová B. 1966. Pollen grains of angiosperms in the Cenomanian Peruc Formation in Bohemia. Palaeobotanist 15:52-54. [ Links ]
Pacltová B. 1981. The evolution and distribution of Normapolles pollen during the Cenophytic. Review of Palaeobotany and Palynology 35:175-208. [ Links ]
Pérez-Lozada M., Høeg J.T. and Crandall K.A. 2004. Unraveling the evolutionary radiation of the Thoracican barnacles using molecular and morphological evidence: a comparison of several divergence time estimation approaches. Systematic Biology 53:244-264. [ Links ]
Phillips M.J. 2009. Branch-length estimation bias misleads molecular dating for a vertebrate mitochondrial phylogeny. Gene 441:132-140. [ Links ]
Phillips T.L. 1980. Stratigraphic and geographic occurrences of permineralized coal-swamp plants - Upper Carboniferous of North America and Europe. In: Dilcher D.L. and Taylor T.N. Eds. Biostratigraphy of Fossil Plants: Successional and Paleoecological Analyses, pp. 25-92, Hutchinson and Ross, Stroudsburg. [ Links ]
Pirie M.D. and Doyle J.A. 2012. Dating clades with fossils and molecules: the case of Annonaceae. Botanical Journal of the Linnean Society 169:84-116. [ Links ]
Posada D. and Buckley T.R. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53:793-808. [ Links ]
Rambaut A. and Grassly N.C. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Computer Applications in the Biosciences 13:235-238. [ Links ]
Ramshaw J.A.M., Richardson D.L., Meatyard B.T., Brown R.H., Richardson M., Thompson E.W. and Boulter D. 1972. The time of origin of the flowering plants determined by using amino acid sequence data of Cytochrome C. New Phytologist 71:773-779. [ Links ]
Rannala B. and Yang Z. 2007. Inferring speciation times under an episodic molecular clock. Systematic Biology 56:453-466. [ Links ]
Rubinstein C.V., Gerrienne P, de la Puente G.S., Astini R.A. and Steemans P. 2010. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytologist 188:365-369. [ Links ]
Rydin C., Källersjö M. and Friis E.M. 2002. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: conflicting data, rooting problems and the monophyly of conifers. International Journal of Plant Sciences 163:197-214. [ Links ]
Sanderson M.J. 1997. A nonparametric approach to estimating divergence times in the absence of rate constancy. Molecular Biology and Evolution 14:1218-1231. [ Links ]
Sanderson M.J. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution 19:101-109. [ Links ]
Sanderson M.J. 2004. r8s version 1.70. r8s software. Analysis of rates of evolution. <http://loco.biosci.arizona.edu/r8s/> (18 May, 2011). [ Links ]
Sanderson M.J. and Doyle J.A. 2001. Sources of error and confidence intervals in estimating the age of angiosperms from rbcL and 18S rDNA data. American Journal of Botany 88:1499-1516. [ Links ]
Sanderson M.J., Wojciechowski M.F., Hu J.-M., Sher Kan T. and Brady S.G. 2000. Error, bias, and long-branch attraction in data for two chloroplast photosystem genes in seed plants. Molecular Biology and Evolution 17:782-797. [ Links ]
Schneider H., Schuettpelz E., Pryer K.M., Cranfill R., Magallón S.and Lupia R. 2004. Ferns diversified in the shadow of angiosperms. Nature 428:553-557. [ Links ]
Scott A. 1974. The earliest conifer. Nature 251:707-708. [ Links ]
Scott A.C. and Chaloner W.G. 1983. The earliest fossil conifer from the Westphalian B of Yorkshire. Proceedings of the Royal Society of London B 220:163-182. [ Links ]
Skog J.E. and Banks H.P 1973. Ibyka amphikoma, gen. et sp. n., a new protoarticulate precursor from the late Middle Devonian of New York State. American Journal of Botany 60:366-380. [ Links ]
Smith S.A. and Donoghue M.J. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322:86-89. [ Links ]
Smith S.A., Beaulieu J.M. and Donoghue M.J. 2010. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proceedings of the National Academy of Sciences, USA 107:5897-5902. [ Links ]
Soltis P.S, Soltis D.E., Savolainen V., Crane P.R. and Barraclough T.G. 2002. Rate heterogeneity among lineages of tracheophytes: Integration of molecular and fossil data and evidence for molecular living fossils. Proceedings of the National Academy of Sciences, USA 99:4430-4435. [ Links ]
Soltis D.E., Smith S.A., Cellinese N., Wurdack K.J., Tank D.C., Brockington, S.F., Refulio-Rodríguez N.F., Walker J.B., Moore M.J., Carlsward B.S., Bell C.D., Latvis M., Crawley S., Black C., Diouf D., Xi Z., Rushworth C.A., Gitzendanner M.A., Sytsma K.J., Qiu Y.L., Hilu K.W., Davis C.C., Sanderson M.J., Beaman R.S., Olmstead R.G., Judd W.S., Donoghue M.J. and Soltis PS. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany 98:704-730. [ Links ]
Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [ Links ]
Stein W.E. Jr. 1982. Iridopteris eriensis from the Middle Devonian of North America, with systematics of apparently related taxa. Botanical Gazette 143:401-416. [ Links ]
Stevens P.F. 2012. Angiosperm Phylogeny Website. Version 12, July 2012. <http://www.mobot.org/MOBOT/research/APweb/> (10 January, 2013). [ Links ]
Swofford D.L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0. Sinauer Associates, Sunderland. [ Links ]
Taylor T.N., Taylor E.L. and Krings M. 2009. Paleobotany. The Biology and Evolution of Fossil Plants, Academic Press, New York. [ Links ]
Thorne J.L. and Kishino H. 2002. Divergence time and evolutionary rate estimation with multilocus data. Systematic Biology 51:689-702. [ Links ]
Thorne J.L., Kishino H. and Painter I.S. 1998. Estimating the rate of evolution of the rate of molecular evolution. Molecular Biology and Evolution 15:1647-1657. [ Links ]
Tims J.D. and Chambers T.C. 1984. Rhyniophytina and Trimerophytina from the early land flora of Victoria, Australia. Palaeontology 27:265-279. [ Links ]
Townrow J.A. 1967. On Rissikia and Mataia podocarpaceous conifers from the lower Mesozoic of southern lands. Proceedings of the Royal Society of Tasmania 101:103-138. [ Links ]
Trevisan L. 1988. Angiospermous pollen (monosulcate-trichoto-mosulcate phase) from the very early Lower Cretaceous of Southern Tuscany (Italy): some aspects. Proceedings 7th International Palynological Congress, Brisbane, Australia, Elsevier, Amsterdam. [ Links ]
Upchurch G.R.Jr., Crane PR. and Drinnan A.N. 1994. The mega-flora from the Quantico locality (upper Albian), Lower Cretaceous Potomac Group of Virginia. Virginia Museum of Natural History Memoir 4:1-57. [ Links ]
von Balthazar M., Pedersen K.R. and Friis E.M. 2005. Teixeiria lusitanica, a new fossil flower from the Early Cretaceous of Portugal with affinities to Ranunculales. Plant Systematics and Evolution 255:55-75. [ Links ]
von Balthazar M., Pedersen K.R., Crane P.R., Stampanoni M. and Friis E.M. 2007. Potomacanthus lobatus gen. et sp. nov., a new flower of probable Lauraceae from the Early Cretaceous (early to middle Albian) of eastern North America. American Journal of Botany 94:2041-2053. [ Links ]
Wertheim J.O., Fourment M., and Kosakovsky Pond S.L. 2012. Inconsistencies in estimating the age of HIV-1 subtypes due to heterotachy. Molecular Biology and Evolution 29:451-456. [ Links ]
Wicke S. and Quandt D. 2009. Universal primers for amplification of the trnK/matK region in land plants. Anales del Jardín Botánico de Madrid 66:285-288. [ Links ]
Wolfe K.H., Guoy M., Yang Y.W., Sharp P.M. and Li W.H. 1989. Date of the monocotdicot divergence estimated from chloroplast DNA sequence data. Proceedings of the National Academy of Sciences USA 86:6201-6205. [ Links ]
Worberg A., Quandt D., Barniske A.M., Löhne C., Hilu K.W. and Borsch T. 2007. Insights from non-coding and rapidly evolving DNA. Organisms Diversity and Evolution 7:55-77. [ Links ]
Yang Z. and Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Molecular Biology and Evolution 23:212-226. [ Links ]
Note added in Proof: As discussed in the manuscript, pollen grains from Valanginian-Hauterivian sediments (ca. 140-130 Ma, Early Cretaceous; Hughes and McDougall, 1987; Hughes et al., 1991; Brenner, 1996) are here interpreted as being very close to the onset of angiosperm crown diversification. Recently reported angiosperm-like pollen grains from Middle Triassic sediments of northern Switzerland (Hotchuli and Feist-Burkhardt. 2013. Frontiers in Plant Science 4:344) are very similar to those from the Early Cretaceous, but are ca. 100 mys older. Considering the absence of a continuous fossil record between these Middle Triassic pollen grains and those from the Early Cretaceous, and in agreement with Hotchuli and Feist-Burkhardt (2013), the former are here interpreted as angiosperm stem lineage members.














