Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Similares em
SciELO
Similares em
SciELO
Compartilhar
Agrociencia
versão On-line ISSN 2521-9766versão impressa ISSN 1405-3195
Agrociencia vol.50 no.8 Texcoco Nov./Dez. 2016
Plant Protection
In vitro activity of Bacillus spp. on mycelial growth inhibition of Fusarium equiseti and Fusarium solani isolated from habanero peppers (Capsicum chinense Jacq.)
1 Instituto Tecnológico de Conkal, km 16.3, Avenida Tecnológico s/n. 97345. Conkal, Yucatán. México. (areyes.itconkal@gmail.com)
Fusarium can induce wilt in habanero peppers (Capsicum chinense Jacq.), it is frequently found in the soil and in various plant species and the use of systemic fungicides is common for it’s control. This disease demands research focused on decreasing the use of fungicides that favor the permanence of resistant populations of plant pathogens and pollute the environment. Antagonists Bacillus species are an alternative for plant pathogen control, such as Fusarium spp., causing fungal etiologies in crops such as the habanero peppers. The objective of this study was to evaluate the in vitro activity of Bacillus spp. on inhibiting mycelial growth of Fusarium equiseti and F. solani, which cause wilting in habanero peppers, in direct confrontations and by using cellfree filtrates to inhibit conidia germination. These allow to detect the presence of the genes responsible for directing the lipopeptides synthesis involved in antifungal activity. Our hypothesis was that at least one strain of Bacillus spp. inhibits F. equiseti and F. solani, and is possible to detect at least one gene for lipopeptid synthesis. The experimental design was completely randomized with four replications. All antagonists inhibited the mycelial growth of F. equiseti (2.15 to 71.55 %) and F. solani (3.76 to 69.16 %). Filtrates of Bacillus subtilis CBRF8 inhibited 100 % conidia germination. Polymerase chain reaction (PCR) amplified three fragments of the genes: bamC, ituA and sfp which correspond to bacillomycin D, iturin A and surfactin.
Keywords: Bacillus; Fusarium; antagonist; phytopathogen; lipopeptides
Fusarium puede inducir marchitez en chile habanero (Capsicum chinense Jacq.), con frecuencia está en el suelo y en especies diversas y el uso de fungicidas sistémicos es común para su control. Esta enfermedad demanda investigación enfocada a disminuir el uso de fungicidas que propician la permanencia de poblaciones resistentes de fitopatógenos y contaminan el ambiente. Las especies de Bacillus antagonistas son una alternativa para el control de fitopatógenos, como Fusarium spp., causantes de etiologías fúngicas en cultivos como el chile habanero. El objetivo de este estudio fue evaluar la actividad in vitro de Bacillus spp. en la inhibición del crecimiento micelial de Fusarium equiseti y F. solani, causantes de la marchitez en chile habanero, en confrontaciones directas y mediante filtrados libres de células para inhibir la germinación de conidios, y detectar la presencia de los genes encargados de dirigir la síntesis de lipopéptidos involucrados con actividad antifúngica. La hipótesis fue que al menos una cepa de Bacillus spp. inhibe a F. equiseti y F. solani, y es posible detectar al menos un gen de la síntesis de lipopéptidos. El diseño experimental fue completamente al azar con cuatro repeticiones. Todos los antagonistas inhibieron el crecimiento micelial de F. equiseti (2.15 a 71.55 %) y F. solani (3.76 a 69.16 %). El filtrado de Bacillus subtilis CBRF8 inhibió 100 % la germinación de los conidios y la Reacción en Cadena de la Polimerasa (PCR) amplificó tres fragmentos de los genes bamC, ituA y sfp, que corresponden a la bacilomicina D, la iturina A y la surfactina.
Palabras clave: Bacillus; Fusarium; antagonista; fitopatógenos; lipopéptidos
Introduction
Phytopathogens such as Fusarium are the main cause of plant wilting. Can also induce root rottening, chlorosis and defoliation (Vásquez López et al., 2009; Villanueva-Arce et al., 2013). To control these pathogens, organosintetic fungicides are frequently applied, which in time, pollute the environment and affect human health and selected resistant fungal strains, thereby reducing their effectiveness (Novaes et al., 2005; Rubio Reque et al., 2008). There are antagonists bacteria in the rhizosphere, able to exert pathogenic control (Guillen-Cruz et al., 2006). This antagonism is partially attributed to the production of a range of secondary metabolites such as bacteriocins, antibiotics, extracellular enzymes (Mojica-Marin et al., 2009), and lipopeptides (iturin, surfactin, fengycin and bacillomycin) (Ramarathnam et al., 2007; Mora et al., 2011; Berić et al., 2012). Bacillus and Paenibacillus species represent a potential to detect metabolites for phytopathogen control, both common and resistant to chemical fungicides, given that they produce these polypeptides (Cochrane and Vederas, 2014). The administration of Bacillus subtilis that release peptides, inhibit the mycelial growth of Fusarium oxysporum and F. solani (Radzhabov and Davranov, 2010). Lipopeptide extracts from B. subtilis and B. amyloliquefaciens showed antifungal activity against F. moniliforme, which produced changes in the hyphae’s cell wall, causing spherical swelling and bumps. These extracts contain compounds related to iturin and fengycin. Protection may function by the secretion of lipopeptides by the rhizobacteria, given that they inhibit pathogens; other mechanisms can also act, like the induction of pathogenesis related genes of host plants, as the lipopeptide extract by itself does not increase gene expression (Gondgenes et al., 2015). In this study, we estimated the antagonistic effectiveness of Bacilli class isolates on two Fusarium species, as these are the cause of wilting in habanero peppers (Capsicum chinense Jacq.), by direct confrontation and conidia germination inhibition by bacterial filtrates. Also, gene activity was analyzed and associated with the biosynthesis of the lipopeptides involved in antifungal activity. Our hypothesis was that a strain of Bacillus spp. has inhibitory activity against F. equiseti and F. solani and has at least one gene for lipopeptide synthesis.
Materials and Methods
Microorganisms used
Sixteen strains of the Bacilli class were used in this study with reported antifungal activity, previously isolated from soil samples from the Yucatan Peninsula, Mexico (Sosa et al., 2012), which belong to the Instituto Tecnológico de Conkal collection, Yucatan. Bacterias were activated on (NA) nutrient agar (BD Bioxon®, Becton Dickinson Mexico). Its purity was verified by growing homogeneous colonies, the presence of Gram positive cells and endospores formation. All strains were grown in the same medium until autolysis at 28 °C for 4 d and stored at 4 °C until they were use.
Isolation and identification of Fusarium spp.
Samples of stems and roots were collected from habanero pepper plants with characteristic symptoms of the disease, in field and greenhouse areas (21° 04’ N and 89° 31’ W) of research and production, at the Instituto Tecnológico de Conkal. Sections of 0.5 cm were disinfected with 2 % sodium hypochlorite for 30 s, washed twice with sterile distilled water, seeded in Petri dishes with growth medium potato dextrose agar (PDA) (BD Bioxon®) and incubated at 28 °C for 72 h, then purified. A preliminarily pathogen identification was done based on their morfotaxonomic characters (Nelson et al, 1983; Barnett and Hunter, 1999).
DNA extractions were taken from monosporic mycelium cultures in order to confirm the specific identification of the fungi (Liu et al., 2002). For this, the ITS1-5.8S-ITS2 rDNA region of was analyzed; primers were those reported by White et al. (1990), ITS1 (5’-TCCGTAGGTGAACCTGCGG-3 ‘) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3’), amplification was performed by PCR (polymerase chain reaction). This included an initial denaturation for 2 min at 95 °C, followed by 30 cycles of denaturation at 94 °C for 1 min, alignment at 54 °C for 30 sand a 1 min extension at 72 °C, and a final 5min extension at 72 °C. The amplified products were visualized via electrophoresis on 1 % agarose gel (SIGMA®, USA) and a 1 kb molecular weight marker (Invitrogen® USA). The amplified PCR products were sequenced at Macrogen (www.macrogenusa.com). The sequences were compared with the GenBank database from the National Center for Biotechnology Information (http:// blast.ncbi.nlm.gov) with the Basic Local Alignment Search Tool (BLAST) program.
Antagonism of bacterial isolates against Fusarium spp.
Bioassays were done in Petri dishes with PDA medium; in the center of each box, 0.5 cm diameter sections of the phytopathogen mycelium and 6 mL of a 1x107 UFC suspension of the bacterial isolates. This was obtained by scraping with a sterile glass slide, from a bacterial culture in AN, after a 5 d incubation period. The suspension was applied at four equidistant points around the fungus, 2 cm apart. The control consisted of Petri dishes with the fungal pathogen but without bacterial strains. The boxes were incubated at 28 °C. After 7 d the mycelial growth inhibition percent (ICR) was determined with the formula proposed by Ezziyyani et al. (2004), also the inhibition halo, between the fungal colony and bacterial strains. Conidia inhibition was determined by the selecting the strains showing a percentage of ICR of at least 65 % and showed an inhibition halo greater at 3.0 mm at least against one of the fungal strains.
Conidia germination inhibition by filtered bacterial
Bacterial isolates were cultured in 250 mL Erlenmeyer flasks, with a 100 mL of Luria-Bertani liquid medium® (LB) (DIBICO®, Mexico), to which 1 mL of bacterial suspension of 1x107 UFC were added. Flasks with the bacterial cells suspension were maintained in an orbital shaker (MaxQ4450, Thermo Scientific, USA) at 200 rpm for 72 h at 29 °C. The supernatant was then recovered by centrifugation at 8765 x g for 10 min, then filtered through Merck® filters (Millipore Germany) of 0.45 μm. and a bacteria free filtrate was obtained. A suspension of 1x108 conidia mL-1 of Fusarium spp. was mixed with the bacteria filtrate (1: 1 v/v). The mixture was seeded in Petri dishes with PDA. Each seed point had a coverslip placed on. The number of germinated conidia after 3, 5, 8 and 10 h was recorded with an optical microscope DM500 (Leica Microsystems, Switzerland) with a 100X objective; values were then converted to percentage. This quantification was suspended when in 100 conidia in random control without filtering, the germ tubes extended over half the length of the size of the fungal cells (Mahadtanapuk et al., 2007; Ruiz-Sanchez et al., 2014).
Identification of isolated bacterial antagonists
The selected Isolates for germination inhibition assays which had previous had a molecular identification were identified. This was done with the analysis of the 16S rDNA gene sequence, and the Eubac27F and Eubac1492R starters (Delong, 1992). To extract the DNA a Wizard Genomic Purification Kit was used (Promega®, USA). The reaction conditions included 1X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs mix, 1.0 mM of each oligonucleotide, 2.0 U Taq DNA polymerase (Invitrogen® USA) and 1mL of DNA at 200ng, to a final 50mL of reaction mixture volume. PCR amplification (in a TECHNE-312, US thermocycler) included initial denaturation at 94 °C for 3 min, followed by 32 cycles (denaturalization at 94 °C for 1 min, 52 °C alignment for 1 min and a 72 °C extension for 1.45 min), final extension at 72 °C for 7 min and 4 °C final temperature. The amplified products and the sequences received the same procedure done with the fungi.
Detection of genes in isolated bacteria for lipopeptide production
The bacterial isolates to detect genes encoding lipopeptid production were the same as those used in the evaluation of the conidia germination inhibition by bacterial filtered, as these were candidates that could produce secondary metabolites with antifungal activity (Athukorala et al.,2009). In order to detect bamC (bacillomycin D), Fend (fengycin), ituA (iturin A), zmaR (zwittermicin A) and sfp (surfactin) genes, the oligonucleotide pairs reported by Ramarathnam et al. (2007) and Athukorala et al. (2009) were used. BACC1 F (GAAGGACACGGCAGAGAGTC) and BACC1 R (CGCTGATGACTGTTCATGCT); FEND1 F (TTTGGCAGCAGGAGAAGTTT) and FEND1 R (GCTGTCCGTTCTGCTTTTTC); ITUD1 F (GATGCGATCTCCTTGGATGT) and ITUD1 R (ATCGTCATGTGCTGCTTGAG); ZWITR2 F (TTGGGAGAATATACAGCTCT) and ZWITR2 R (GACCTTTTGAAATGGGCGTA), SUR3 F (ACAGTATGGAGGCATGGTC) SUR3 R (TTCCGCCACTTTTTCAGTTT) (Invitrogen®, USA). The PCR program BACC1 F / R consisted on a denaturation at 94 °C for 3 min, 35 cycles of denaturation at 94 °C for 1 min, alignment at 60 °C for 30 s, extension at 72 °C for 1.45 min and final extension at 72 °C for 6 min (Ramarathnam et al., 2007); for ITUD1F / R and SUR3 F / R consisted of denaturation at 94 °C for 15 min, 30 denaturation cycles at 94 °C for 30 s, alignment at 60 °C for 30 s, extension at 72 °C for 2 min and a final extension at 72 °C for 10 min (Stankovic et al., 2012.); for FEND1 F/R consisted of denaturation at 94 °C for 3 min, 45 cycles of denaturation at 94 °C for 1 min, alignment at 62 °C for 1 min, extension at 72 °C for 1.45 min and final extension at 72 °C for 6 min (Ramarathnam et al., 2007) and for ZWTR2 F / R consisted of denaturation at 94 °C for 3 min, followed by 30 denaturation cycles at 94 °C for 15 s, alignment at 60 °C for 45 s, extension at 72 °C for 2 min and final extension at 72 °C for 4 min (Basurto et al., 2012).
Data analysis
The experimental design was completely randomized and treatments had four repetitions. An ANOVA was performed on the data, with a previous data transformation to arcsin [y=arcsin percentage (sqrt x/100)], and means were compared with the Tukey test (p£0.05). Data was analyzed with SAS Ver. 9.1 (SAS Institute Inc. 2010).
Results and Discussion
Isolation and identification of fungal strains
Two Fusarium species were isolated from Capsicum chinense Jacq. plants with necrosis symptoms in stems and roots. One was identified as F. equiseti with ITCF1 registration, which had abundant, aerial cottony white mycelium, with rapid growth and orange color on its underside. Microscopic examination revealed the presence of abundant macroconidia with attenuated apical cells, of one to four cells. Microconidia generally present, usually less than simple cells, oval or kidneyshaped produced in false heads. The other isolated species was identified as F. solani with ITCF2 registration. It had sparse aerial mycelium, relatively slower growth, and cream color on both sides. Microscopic examination revealed the presence of few cylindrical macroconidia, round simple basal and apical cells or single cell; abundant microconidia, generally single cell, and in some cases cylindrical and basal attenuated apical foot shaped cells (Nelson et al., 1983; Escalona et al., 2006). The analysis of the ITS1-5.8s-ITS2 rDNA region confirmed a 100 % similarity of the ITCF1 strain with F. equiseti (GenBank JQ690081) and in ITCF2 case, the similarity was of 99 % regard F. solani (JQ910159). These are important plant pathogens that cause losses in crop production, such as C. annuum. (Martinez et al., 2011;Sanzon Gomez et al., 2012), wheat (Triticum aestivum L.), soybean (Glycine max L.) and pea (Pisum sativum L.) (Ivić et al., 2009; Espinoza et al., 2011), corn (Zea mays L.) (Zainudin et al., 2011), tomato (Solanum lycopersicum L.) and squash (Cucurbita pepo L.) (Montano et al., 2012; Roberti et al., 2012).
Bacterial isolates antagonistic activity against F. Equiseti and F. Solani
Fusarium spp. ICR. The analysis of variance for the ICR variable showed significant differences between the 16 bacterial isolates (p≤0.05). ICR intervals in both species of Fusarium were from 2.15 to 71.55 %. In F. equiseti ITCF1 bacterial isolates with CBRF8, CBCK41, CBMT51 and CBRM17 registration were those who induced the highest averages of ICR percentage, with 65.70, 66.15, 71.23 and 71.55 % (p£0.05). In F.solani ITCF2 the CBRF15, CBRF9 and CBRF8 isolates outperformed on average the other bacterial isolates (p≤0.05), given that they caused an ICR of 61.60, 62.67 and 69.16 % (Table 1). The antifungal activity of the bacterial isolates become clear in Fusarium sp. when it was confronted with B.subtilis LSB4 and Bacillus sp. LSB9, as they showed mycelial growth inhibition of 90 and 90.4 % each (Badia et al., 2011). Inhibitions in those studies exceeded our results. However, in other cases, the effectiveness with F. oxysporum was similar to that estimated for isolated CBRF8, CBRM17 and CBMT51 (Li et al., 2012b), or less effective, although it came to similar or different species of the fungi (Romano et al., 2013; Sarti and Miyazaki, 2013) and even when evaluated on other Fusarium strains. Antagonism response is associated with the synthesis of lipopeptides like iturin, surfactin, fengicyn and bacillomycin (Ramarathnam et al., 2007; Mora et al., 2011; Berić et al., 2012) and secondary metabolites that can generate an inner lysis area and a thickening edge in the inhibition zone. This effect was observed in F. equiseti and F. solani. They also affected the radial growth of the fungi, as it happened against Curvularia lunata (Basha y Ulaganathan, 2002; Orberá et al., 2009).
Table 1 Growth inhibition and inhibition halos by action of bacterial isolates against strains of Fusarium equiseti and F. solani.
| Aislados | F. equiseti ITCF1 | F. solani ITCF2 | ||||
| ICR (%) | Halos (mm) | ICR (%) | Halos (mm) | |||
| CBRF8 | 65.70 ± 3.65 ab | 0.00 ± 0.0 e | 69.16 ± 1.77 a | 0.00 ± 0.0 b | ||
| CBRF9 | 54.85 ± 2.84 hijk | 0.00 ± 0.0 e | 62.67 ± 6.07 ab | 0.00 ± 0.0 b | ||
| CBRF12 | 60.54 ± 2.59 gh | 0.00 ± 0.0 e | 58.73 ± 0.60 bcde | 0.00 ± 0.0 b | ||
| CBRF15 | 58.24 ± 1.82 hi | 1.49 ± 0.17 d | 61.60 ± 1.57 bc | 1.30 ± 0.21 a | ||
| CBRF24 | 57.75 ± 4.72 hi | 1.02 ± 0.72 d | 58.25 ± 2.44 bcdef | 0.00 ± 0.0 b | ||
| CBRM17 | 71.55 ± 0.71 a | 5.72 ± 0.32 a | 52.74 ± 2.45 defg | 0.00 ± 0.0 b | ||
| CBMT2 | 64.26 ± 2.00 bc | 3.52 ± 0.51 b | 52.60 ± 1.82 efg | 0.00 ± 0.0 b | ||
| CBMT51 | 71.23 ± 1.18 a | 6.10 ± 0.67 a | 55.02 ± 4.42 cdef | 0.00 ± 0.0 b | ||
| CBCC2 | 48.76 ± 3.66 g | 0.00 ± 0.0 e | 53.66 ± 2.16 defg | 0.00 ± 0.0 b | ||
| CBDG60 | 60.71 ± 1.35 bcd | 0.00 ± 0.0 e | 59.58 ± 1.24 bcd | 0.00 ± 0.0 b | ||
| CBSN67 | 53.35 ± 1.90 efg | 0.00 ± 0.0 e | 55.45 ± 2.85 cdef | 0.00 ± 0.0 b | ||
| CBMN22 | 50.60 ± 4.30 fg | 0.00 ± 0.0 e | 51.58 ± 1.26 fg | 0.00 ± 0.0 b | ||
| CBCK36 | 51.14 ± 1.14 fg | 0.00 ± 0.0 e | 47.81 ± 1.63 g | 0.00 ± 0.0 b | ||
| CBCK41 | 66.15 ± 0.88 ab | 2.39 ± 0.37 c | 20.19 ± 3.43 h | 0.00 ± 0.0 b | ||
| CBCK46 | 57.00 ± 3.29 def | 0.00 ± 0.0 e | 3.76 ± 2.71 i | 0.00 ± 0.0 b | ||
| CBCK47 | 2.15 ± 0.77 h | 0.00 ± 0.0 e | 57.24 ± 3.01 bcdef | 0.00 ± 0.0 b | ||
| Testigo | 0.00 ± 0.0 h | 0.00 ± 0.0 e | 0.00 ± 0.0 i | 0.00 ± 0.0 b | ||
| DMS | 6.55 | 0.76 | 6.96 | 0.12 | ||
Means with different letter are statistically significant (p ≤ 0.05). ICR: inhibition of radial growth; LSD: Least significant difference.
Presence and size of inhibition halo
The analysis of variance of the inhibition halos showed significant differences between the 16 bacterial isolates (p≤0.05). Halos generated in both Fusarium species were of 1.30 to 6.10 mm. In F. equiseti ITCF1 the isolates CBRM17 and CBMT51 were different to the rest of the isolates (p≤0.05), because they generated the largest inhibition halos (5.72 and 6.10 mm). In F. solani ITCF2 only the CBRF15 isolate generated an inhibition halo of 1.30 mm, but it was not the bacteria with the greatest inhibition of mycelial growth (Table 1). The formation of the inhibition halos in bacterial isolates has been evident in the direct contrast of B. cereus X16 against F. roseum (Sadfi et al., 2002). The growth inhibition halos due to the CBRM17 and CBMT51 isolates in F. equiseti showed whitish appearance, with quadrangular delimitation of the fungus. This activity was also observed in Colletotrichum gloeosporioides, by action of B. subtilis and B. licheniformis (Gutierrez-Alonso et al., 2003). The areas bounded by isolates against fungi are due to antifungal metabolites production, which are diffusible in the culture medium (Sadfi et al., 2002). These antifungal metabolites may be cyclic lipopeptides, as surfactin, iturin A and fengicyn, acting as biocontrol agents against Fusarium (Arguelles-Arias et al., 2009).
Inhibition on conidia germination by filtered bacteria
CBRM17, CBMT51 and CBMT2 isolates met the criteria set for this test. The average inhibition halo of F. equiseti exceeded 3.0 mm. But the CBRF8 isolate was determined because of its ICR activity in both phytopathogenic species. Bacterial filtered inhibited between 42.25 and 100 % conidia germination of F. equiseti, and for F. solani the averages were from 12.75 to 100 % (Table 2). The filtering with the highest antifungal capacity was that of the isolated CBRF8, for it inhibited a 100 % of the conidia germination of both fungus species (Figure 1). The 100 % effectiveness was also observed on Colletotrichum musae, exposed to filtered B. licheniformis, B. amyloliquefaciens and B. subtilis; although, it formed swellings and distortions in the conidia and germ tubes of the fungus. Similar activity was not observed in that study (Mahadtanapuk et al., 2007). CBRF8 isolated filtrates are candidates for antifungal activity control, exceeding by 40 % the effectiveness of filtered B. subtilis applied to Aspergillus flavus conidia (Zhang et al., 2010), and 63 % to those from Bacillus sp. on C. gloesporioides conidia (Gutierrez-Alonso et al., 2003). The CBRF8 bacterial isolate did not showed inhibition halos in the tested fungal strains, but the antifungal capacity of the filtered may be associated to the production of proteins with enzymatic or toxic activity, which together cause somatic structural changes on the fungi and in the conidia viability (Basurto-Cadena et al., 2010). These can affect the overall growth of the fungi or cause only morphological changes in the mycelium or in its resistance and reproduction structures (Carissimi et al., 2009).
Table 2 Conidia germination inhibition (%) of Fusarium equiseti and F. solani by bacterial filtered after 10 h exposure.
| Aislados | F. equiseti ITCF1 | F. solani ITCF2 |
| CBRF8 | 100 ± 0.0 a† | 100 ± 0.0 a |
| CBRM17 | 69.50 ± 5.80 b | 3.50 ± 1.00 d |
| CBMT2 | 49.25 ± 5.62 c | 12.75 ± 2.06 c |
| CBMT51 | 42.25 ± 9.03 c | 15.50 ± 0.58 b |
| Testigo | 0.00 ± 0.0 d | 0.00 ± 0.0 e |
| DMS | 11.83 | 2.30 |
Means with different letters are statistically significant (p≤0.05). LSD: Least significant difference.
Identification of two isolated bacterial antagonists
CBMT2 and CBMT51 isolates were identified as B. subtilis (Ruiz t al., 2016); so, in our study the CBRM17 and CBRF8 isolates were analyzed, based on the 16S rDNA gene sequence. The amplified PCR product was about 1500 bp, and had a 94 to 99 % similarity with the CBRF8 isolated with Bacillus subtilis (GQ214132.1) and CBRM17 with Paenibacillus sp. (KJ948328.1). Although these isolates are of different genera, they belong to the same Bacilli class. There is documented diversity of Bacillus antagonists, and B. subtilis is the one that has the most recognized antifungal properties against Fusarium (Guillen-Cruz et al., 2006; Badía et al., 2011; Li et al., 2012a). Also Paenibacillus polymyxa, P. peoriae, P. brasilensis, P. alginolyticus and P. favisporus show antifungal activity against F. oxysporum (Li et al., 2012b; Sato et al., 2014).
Detection of genes encoding antifungal lipopeptides biosynthesis
Amplified specific products were only observed in the CBRF8 bacterial isolate with the expected size for the bamC, ituA and sfp gene fragments of 875, 675 and 500 bp, each (Ramarathnam et al., 2007; Berić et al., 2012; Stanković et al., 2012.) (Figure 2). In the CMBR17, CBMT51 and CBMT2 isolates there was no detection of these fragments. The detected genes in the isolated CBRF8 coded bacillomycin D production, iturin A and surfactin. The presence of more than one gene for lipopeptides synthesis in the B. subtilis CBRF8 isolate, is a characteristic of the isolates of this genere, in which there has been obtained 167 isolates having the bacillomycin D gene, 111 the surfactin gene and 79 the iturin gene (Stankovic et al., 2012). In same and different species the bacillomicyn genes presence has been detected, as in B. cereus DFE4, B amyloliquefaciens DEF16 and BS6 and B. subtilis 49, the fengycin gene in B. subtilis DFH08 and 49; although there was no amplification for this gene, its synthesis is associated with 60 % growth inhibition of F. graminearum and Sclerotinia sclerotiorum (Ramarathnam et al., 2007). This species presents iturin C and D genes and bacillomicin C and AB, related to radial growth inhibition, inhibition halos and inhibition of conidial germination in F. oxysporum and F. solani (Chung et al., 2008). These mechanisms of lipopeptide synthesis genes with antifungal metabolites or compounds of various peptide and lipopeptide nature, such as iturins, fengycins and surfactins, can interfere with the antifungal activity of B. subtilis against phytopathogens originated in the ground (Cazorla et al., 2007).
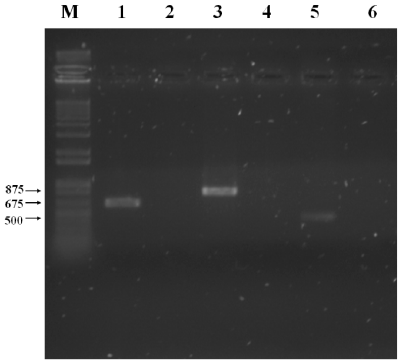
Figure 2 PCR amplification of the fragments of genes for lipopeptides biosynthesis from CBRF8 isolated, with the specific oligonucleotides. Lane M, molecular weight marker (1 kb DNA Ladder; Invitrogen ® ); lane 1, ITUD1; lane 2, FEND1; lane 3, BACC1; lane 4, ZWITR2; lane 5 SUR3; lane 6 negative control.
Conclusions
Fusarium equiseti and F. solani species were the causative wilting agents in C. chinense. In the fungi mycelial growth inhibition, the best in vitro antagonism against F. equiseti was exerted by Paenibacillus sp. CBRM17, B. subtilis CBMT51 and CBRF8 isolates, with 71.55, 71.23 and 65.70 %. Against F. solani, the highest antagonism was estimated with B. subtilis CBRF8 and Bacillus sp. isolated. CBRF9 with 69.16 and 62.67 %. Only the B. subtilis CBRF8 isolate caused 100 % inhibition of conidial germination. The presence of genes encoding the synthesis of lipopeptides was detected only with the B. subtilis CBRF8 isolated, which presented activity with, bamC, ituA and sfp genes responsible for the synthesis of bacillomicin D, iturin A and surfactin, respectively.
Literatura citada
Arguelles-Arias, A., M. Ongena, B. Halimi, Y. Lara, A. Brans, B. Joris, and P. Fickers. 2009. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 8: 1-12. [ Links ]
Athukorala, S. N., W. G. Dilantha F., and K. Y. Rashid. 2009. Identification of antifungal antibiotics of Bacillus species isolated from different microhabitats using polymerase chain reaction and MALDI-TOF mass spectrometry. Can. J. Microbiol. 55: 1021-1032. [ Links ]
Badía, M. R., B. Hernández T., J. A. Murrel L., J. Mahillon, y M. Pérez H. 2011. Aislamiento y caracterización de cepas de Bacillus asociadas al cultivo del arroz (Oryza sativa L.). Rev. Bras. Agroecol. 6: 90-99. [ Links ]
Barnett, H. L., and B. B. Hunter. 1999. Illustrate Genera of Imperfect Fungi, Fourth Edition. Burgess Publishing Company. Minnesota. pp. 118-132. [ Links ]
Basha S., and K. Ulaganathan. 2002. Antagonism of Bacillus species (strain BC121) towards Curvularia lunata. Curr. Sci. 82: 1457-1463. [ Links ]
Basurto-Cadena, M. G., M. I. Font San Ambrosio, J. García Jiménez, y M. Vázquez-Arista. 2010. Cambios en la estructura celular durante la actividad antagónica de Bacillus subtilis contra Rhizoctonia Solani y Fusarium verticillioides. Acta Microscópica 19: 138-144. [ Links ]
Berić, T., M. Kojić, S. Stanković, L. Topisirović, G. Degrassi, M. Myers, and D. Fira. 2012. Antimicrobial activity of Bacillus sp. natural isolates and their potential use in the biocontrol of phytopathogenic bacteria. Food Technol. Biotechnol 50:25-31. [ Links ]
Carissimi, M., G. M. Schipani, J. C. Germani, and S. T. Van Der Sand. 2009. Antifungal activity of Bacillus sp. E164 against Bipolaris sorokiniana. Biociências 17: 48-58. [ Links ]
Cazorla, F. M., D. Romero, A. Pérez-García, B. J. Lugtenberg J., A. D. Vicente, and G. Bloemberg. 2007. Isolation and characterization of antagonistic Bacillus subtilis strains from the avocado rhizoplane displaying biocontrol activity. J. Appl. Microbiol. 103: 1950-1959. [ Links ]
Cochrane, S. A. and J. C. Vederas. 2014. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 36: 4-31. [ Links ]
Chung, S., H. Kong, J. S. Buyer, D. K. Lakshman, J. Lydon, S. D. Kim, and D. P. Roberts. 2008. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soil borne pathogens of cucumber and pepper. Appl. Microbiol. Biotechnol. 80:115-123. [ Links ]
Delong E. F. 1992. Archaea in Coastal Marine Environments. Proc. Natl. Acad. Sci. USA. pp: 5685-5698. [ Links ]
Escalona, Y., D. Rodríguez, y N. Contreras. 2006. Patógenos del suelo en el cultivo de pimentón en la zona baja del municipio Jiménez estado Lara, Venezuela. Rev. Bioagro 18: 3-13. [ Links ]
Espinoza, M. E. B., S. G. Mir L., y M. Amaro. 2011. Hongos asociados al grano de trigo sembrado en áreas del centro de México. Rev. Mex. Fitopatol. 29: 175-177. [ Links ]
Ezziyyani, M., C. Pérez S., M. E. Requena, L. Rubio, y M. E. Candela. 2004. Biocontrol por “ Streptomyces rochei” Ziyani -, de la podredumbre del pimiento (“Capsicum annuum L.”) causada por “Phytophthora capsici”. An. Biol. 26: 69-78. [ Links ]
Gond, S. K., M. S. Bergen, M. S Torres, and J. F. White, 2015. Endophytic Bacillus spp. produces antifungal lipopeptides and induces host defense gene expression in maize. Microbiol. Res. 172: 79-87. [ Links ]
Guillén-Cruz, R., F. D. Hernández-Castillo, G. Gallegos-Morales, R. Rodríguez-Herrera, C. N. Aguilar-González, E. Padrón-Corral, and M. H. Reyes-Valdés. 2006. Bacillus spp. as biocontrol in an infested soil with Fusarium spp., Rhizoctonia solani Kühn, and Phytophthora capsici Leonian, and its effect on development and yield of pepper crop (Capsicum annuum L.). Rev. Mex. Fitopatol. 24: 105-114. [ Links ]
Gutiérrez-Alonso, J. G., O. Gutiérrez A., D. Nieto A., D. Téliz O., E. Zavaleta-Mejía, F. Delgadillo S., y H. Vaquera H. 2003. Evaluación in vitro de Agentes Biológicos y Físicos para el Control de Colletotrichum gloeosporioides (Penz.) Penz. y Sacc. Rev. Mex. Fitopatol. 21: 199-206. [ Links ]
Ivić, D., A. M. Domijan, M. Peraica, T. Miličević, and B. Cvjetković. 2009. Fusarium spp. contamination of wheat, maize, soybean, and pea grain in Croatia. Arch. Ind. Hyg. Toxicol. 60: 435-442. [ Links ]
Li, H., X. Wang, M. Han, Z. Zhao, M. Wang, Q. Tang, C. Liu, B. Kemp, Y. Gu, S. Shuang, and Y. Xue. 2012a. Endophytic Bacillus subtilis ZZ120 and its potential application in control of replant diseases. Afr. J. Biotechnol. 11: 231-242. [ Links ]
Li, P., L. Ma, Y. L. Feng, M. H. Mo, F. X. Yang, H. F.Dai, and Y. X. Zhao. 2012b. Diversity and chemotaxis of soil bacteria with antifungal activity against Fusarium wilt of banana. J. Ind. Microbiol. Biotechnol. 39: 1495-1505. [ Links ]
Liu, D., L. Pearce, G. Lilley, S. Coloe, R. Baird, and J. Pedersen. 2002. PCR identification of dermatophyte fungi Trichophyton rubrum, T. soudanense and T. gourvilii. J. Med. Microbiol. 51: 117-122. [ Links ]
Mahadtanapuk, S., M. Sanguansermsri, R. W. Cutler, V. Sardsud, and S. Anuntalabhochai S. 2007. Control of anthracnose caused by Colletotrichum musae on Curcuma alismatifolia Gagnep. using antagonistic Bacillus spp. Am. J. Agric. Biol. Sci. 2: 54-61. [ Links ]
Martínez, M. A., M. C. Martínez, P. Bielza, J. Tello, and A. Lacasa. 2011. Effect of biofumigation with manure amendments and repeated biosolarization on Fusarium densities in pepper crops. J. Ind. Microbiol. Biotechnol. 38: 3-11. [ Links ]
Mojica-Marín, V., H. A. Luna-Olvera, C. F. Sandoval-Coronado, B. Pereyra-Alférez, L. H. Morales-Ramos, N. A. González Aguilar, y O. G. Alvarado-Gómez. 2009. Control biológico de la marchitez del chile (Capsicum annuum L.) por Bacillus thuringiensis. Phyton. Rev. Int. Bot. Exp. 78: 105-110. [ Links ]
Mora, I., J. Cabrefiga, and E. Montesinos. 2011. Antimicrobial peptide genes in Bacillus strains from plant environments. Int. Microbiol. 14: 213-223. [ Links ]
Montano, M., Y. Duarte, and B. Martínez. 2012. Fusarium species identified from soil and tomato (Solanum lycopersicum L.) in Granma, Cuba. Rev. Prot. Veg. 27:213. [ Links ]
Nelson, P. E., T. A. Toussoun, and W. F. Marasas. 1983. Fusarium species: An Illustrated Manual for Identification. Edit. The Pennsylvania State University. pp. 85-95, 143-150. [ Links ]
Novaes, S., M. De Resende, E P. Paiva. 2005. Efeito de diferentes fungicidas e de resistencia na inibicao do crescimento mivelial de Fusarium oxysporum f.p. gladioli e comparacao de metodologias de inoculacao em bulbos. Sum. Phytopathol. 31: 371-373. [ Links ]
Orberá, R. T. M., M. J. Serrat D., y Z. González G. 2009. Potencialidades de bacterias aerobias formadoras de endosporas para el biocontrol en plantas ornamentales. Rev. Fitosanidad 13: 95-100. [ Links ]
Radzhabov, U. R., and K. Davranov. 2010. Metabolites of Bacillus subtilis SKB 256, growth inhibitors of phytopathogenic fungi. Chem. Nat. Compd. 46: 160-162. [ Links ]
Ramarathnam, R., S. Bo, Y. Chen, W. G. Fernando, G. Xuewen, and T. de Kievit. 2007. Molecular and biochemical detection of fengycin-and bacillomycin D-producing Bacillus spp., antagonistic to fungal pathogens of canola and wheat. Can. J. Microbiol. 53:901-911. [ Links ]
Roberti, R., A. Veronesi, and F. Flamigni. 2012. Evaluation of microbial products for the control of zucchini foot and root rot caused by Fusarium solani f. sp. cucurbitae race 1. Phytopathol. Medit. 51:317-331. [ Links ]
Romano, A., D. Vitullo, M. Senatore, G. Lima and V. Lanzotti. 2013. Antifungal Cyclic Lipopeptides from Bacillus amyloliquefaciens Strain BO5A. J. Nat. Prod. 76: 2019-2025. [ Links ]
Rubio-Reque, G. L., F. M. Baltodano-Sánchez, L. I. Abanto-Campos, J. H. Wilson-Krugg, and M. A. Muñoz-Ríos. 2008. Resistencia in vitro de Rhizoctonia solani y Fusarium oxysporum a los fungicidas Benzomil 500, Rhizolex-T y Homai-WP. Rebiol 28: 1-12. [ Links ]
Ruiz-Sánchez, E., M. A. Mejía-Bautista, A. Serrato-Díaz, A. Reyes-Ramírez, Y. Estrada-Girón, and A. J. Valencia-Botín. 2016. Antifungal activity and molecular identification of native strains of Bacillus subtilis. Agrociencia 50: 133-148. [ Links ]
Ruiz-Sánchez, E., M. A. Mejía-Bautista, J. Cristóbal-Alejo, A. Valencia-Botín, y A. Reyes-Ramírez. 2014. Actividad antagónica de filtrados de Bacillus subtilis contra Colletotrichum gloeosporioides (Penz.). Rev. Mex. Cien. Agric. 5: 1325-1332. [ Links ]
Sadfi, N., M. Cherif, M. R. Hajlaoui, A. Boudabbous, and R. Belanger. 2002. Isolation and partial purification of antifungal metabolites produced by Bacillus cereus. Ann. Microbiol. 52: 323-338. [ Links ]
Sanzón-Gómez, D., G. Valdovinos-Ponce, R. I. Rojas-Martínez, E. Zavaleta-Mejía, M. Avilés, and L. Guevara-Olvera. 2012. Morphological changes in cells of chilli CM334 inoculated with Phytophthora capsici and Fusarium oxysporum. Rev. Mex. Fitopatol. 30: 66-71. [ Links ]
SAS Institute. 2010. User’s Guide: Statistics, version 9.1. SAS Inst. Inc., Cary, North Caroline, USA. [ Links ]
Sato, I., S. Yoshida, Y. Iwamoto, M. Aino, M. Hyakumachi, M. Shimizu, and S. Tsushima. 2014. Suppressive potential of Paenibacillus strains isolated from the tomato phyllosphere against Fusarium crown and root rot of tomato. Microbes Environ. 29: 168-177. [ Links ]
Sarti, G., C y S. S. Miyazaki. 2013 Actividad antifúngica de extractos crudos de Bacillus subtilis contra fitopatógenos de soja (Glycine max) y efecto de su coinoculación con Bradyrhizobium japonicum. Agrociencia 47: 373-383. [ Links ]
Sosa-Pech, M., E. Ruiz-Sánchez, M. Mejía-Bautista, A. Reyes Ramírez, J. Cristóbal-Alejo, A. Valencia-Botín, y O. Gutiérrez-Alonzo. 2012. Actividad antagónista in vitro de aislados de la clase Bacilli de la Península de Yucatán contra cuatro hongos fitopatógenos. Universidad y Ciencia 28: 279-284. [ Links ]
Stanković, S., S. Mihajlović, V. Draganić, I. Dimkić, G. Vukotić, T. Berić, and D. Fira. 2012. Screening for the presence of biosynthetic genes for antimicrobial lipopeptides in natural isolates of Bacillus sp. Arch. Biol. Sci. 64: 1425-1432. [ Links ]
Villanueva-Arce, R., C. A. Aguilar-Pompa, Y. de las M. Gómez-Gómez, G. Valencia-Toro, A. B. Piña-Guzmán, y S. Bautista-Baños. 2013. Control de bacterias patógenas y hongos de postcosecha con extractos del pigmento de Gibberella zeae (Fusarium graminearum). Agrociencia 47: 691-705. [ Links ]
Vásquez-López, A., B. Tlapal-Bolaños, M. J. Yáñez-Morales, R. Pérez-Pacheco, y M. Quintos-Escalante. 2009. Etiología de la marchitez del ‘chile de agua’ (Capsicum annuum L.) en Oaxaca, México. Rev. Fitotec. Mex. 32: 127-134. [ Links ]
White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications. Innis, M. A., D. H. Gelfand, J. J. Sninsky, and T. J. White. (eds). Academic Press, San Diego, CA. pp. 315-321. [ Links ]
Zainudin, N. A. I. M., S. Nordahliawate, N. A. Johari, A. A Razak, and B. Salleh. 2011. Isolation and identification of Fusarium species associated with Fusarium ear rot disease of corn. Pertanika. J. Trop. Agric. Sci. 34: 325-330. [ Links ]
Zhang, D. J., R. F. Liu, Y. G. Li, L. M. Tao, and L. Tian. 2010. Two new antifungal cyclic lipopeptides from Bacillus marinus B-9987. Chem. Pharm. Bull. 58: 1630-1634. [ Links ]
Received: October 2015; Accepted: July 2016











 texto em
texto em 



