Introduction
The receptor for advanced glycation end products (RAGE) binds multiple ligands, including advanced glycation end products (AGEs) [1,2], the high mobility group box-1 protein (HMGB1) [3-5], several members of the S100 calcium-binding protein family, notably S100B [6-9], and the amyloid β peptide [10,11], among several others, which are expressed in inflammatory disorders [6,12-14]. Several studies exploring this concept have been carried out in in vitro conditions with human cells from the immune system, as well as in murine models. These findings include the activation of the membrane surface RAGE receptor upon binding the DNA-HMGB1 complex, HMGB1 or S100B [15]. These interactions induce the expression of pro-inflammatory molecules such as interleukin 8 (IL8), RAGE, HMGB1, and the S100 protein genes [16,17]. In different disease models, such as lupus, diabetes, viral infections and cancer, changes in the levels of soluble RAGE (sRAGE) and membrane RAGE have been observed, suggesting a major role for this protein in the amplification of inflammatory responses [18]. Leukocytes have been the most frequently used cell preparation to study cell receptor-ligand interactions at the membrane level [19]. However, more recently, other cell types have been studied in inflammatory microenvironments, including epithelial and endothelial cells, fibroblasts, and other cells that can respond to various inflammatory stimuli or cell damage. The endothelial cells of veins and arteries can be in direct contact with these pro-inflammatory ligands through RAGE and other receptors, such as Toll-like or cytokine receptors, to activate the MAPK pathways. In turn, these kinases can regulate transcription factors linked to inflammation. The nuclear factor kappa B (NFκB) requires kinase activation to amplify the response to injury or infection. This knowledge is derived from observations of in vitro models that primarily use specific inhibitors or antibodies that block the binding of membrane receptors to their ligands on the cell surface, thus emphasizing the importance of these interactions for cell responses. For instance, in 2003, Treutiger et al. [20] demonstrated that the use of anti-RAGE antibodies reduced the adhesion of neutrophils to human umbilical vein endothelial cells (HUVEC’s) via NFκB signaling in inflammatory conditions elicited by lipopolysaccharide (LPS) or tumor necrosis factor-alpha (TNF-α). Similar results were obtained when HUVEC’s were pre-activated using AGE-albumin induction and exposed to proteins of the S100 family as RAGE ligands [6]. Affinity studies of the S100 family as RAGE ligands have also been reported using surface plasmon resonance (SPR) analysis, and a stoichiometry of 2:1 (S100:RAGE) was determined [7,8,21,22]. These studies also demonstrated that these interactions induce pro-inflammatory responses in cells (including HUVEC’s and macrophages); however, the binding of RAGE to its ligands remains unverified at the cytochemical level. Therefore, in this study, we characterized the early co-localization of endogenous RAGE with endogenous S100B in HUVEC’s challenged with the pro-inflammatory stimulus LPS. In order to confirm the co-localization of these proteins and their interactions. Since, it is known that the activation of RAGE induced by S100B constitutes an important pro-inflammatory pathway during the progression of acute and chronic inflammatory diseases [7,23,24], our study provides insights into the co-localization of these proteins in unstimulated and LPS-stimulated HUVEC’s. Our results represent a basis for ongoing studies involving the entire RAGE-S100B complex and for novel approaches to interfere with inflammatory responses.
Experimental
Reagents
Primary antibodies against RAGE (Ab3611 for immunofluorescence assays against the V and C2 domains) and secondary antibodies were purchased from Abcam (Cambridge, MA, USA). The antibody recognizing S-100B (AB11178) was also purchased from Abcam.
Cell cultures and experimental treatments
Endothelial cells from the veins of human umbilical cords (HUVEC’s) were obtained from eutocic births and caesarean sections, according to the procedure described by Jaffe [25]. Samples were obtained at the Hospital of Obstetrics and Gynecology “Dr. Luis Ayala Castelazo”, IMSS, Mexico City, following a clinical protocol previously approved by the Ethics Committee of the Hospital. Cords were processed within the first 3 h after being obtained, washed and cleaned with sterile saline solution. Once collected, the veins were cut into 5 cm pieces in order to use samples of the same length. The veins were placed in containers with phosphate buffered saline (PBS) supplemented with antibiotic-antimycotic (penicillin/streptomycin 1.5%; GIBCO, Life Technologies, USA plus Amphotericin B, Biowest S.A.S., USA). The vascular endothelium was disaggregated enzymatically by adding 10 ml of 0.02 % collagenase (type 4, Worthington Biochemical Corporation) dissolved in PBS. Endothelial cells were dissociated and collected in fetal bovine serum (FBS, Invitrogen, Mexico). The cells obtained after enzymatic digestion were centrifuged at 800 ( g for 5 min in a Labofuge 400 Heraeus Instruments centrifuge (Thermo Scientific, Waltham, Massachusetts, USA) using a rotor model 8179. The pellets were resuspended in M199 medium (GIBCO, Rockville, Maryland, USA) supplemented with 1 % glutamine (GIBCO), 0.1 mg/ml of endothelial growth factor (Biomedical Technologies Inc., Alfa Aesar, Ward Hill, Massachusetts, USA), 0.1 mg/ml of heparin (Sigma-Aldrich Co., St. Louis, Missouri, USA) and 10 % FBS. Streptomycin and penicillin were added in a 1 % dilution (GIBCO). All cells were maintained by incubation at 37° C with controlled relative humidity and 5 % CO2. For experimental purposes, cells were dissociated from the culture dish with a trypsin/versene solution (GIBCO) for 2 min and reseeded in 48 or 6 wheel-plates at densities ranging from 30,000 to 50,000 cells per wheel for the 48 wheel-plates, or 250,000 for the 6 wheel-plates, in M-199 medium supplemented as previously mentioned. All experiments were performed 24 h later.
The assay for inflammatory activation is based on previous studies under in vitro and ex vivo conditions in HUVEC’s and human umbilical veins stimulated with TNF-α [26]. In these cases, it is known that concentrations of 1-10 ng/ml during 2-6 hours of exposure induce the expression of adhesion molecules and pro-inflammatory cytokines that promote leukocyte adhesion in response to TNF-α. The concentration of LPS that we used in this study is the most commonly used in studies of leukocyte activation involving 2-12 h periods of exposure [27]. Endothelial cell activation was determined after induction with LPS (10 ng/ml). TNF-α (5 ng/ml) was employed in additional experiments as a positive control of inflammation; this concentration was determined from a concentration curve in a leukocyte/endothelial cell adhesion assay, ranging from 0.5 to 10 ng/ml. Endothelial cells were washed twice in PBS and placed in new culture medium with 10 ng/ml of TNF-α or LPS for 3 hours at 37 °C, 5% of CO2 and 95% air. Immediately thereafter, all samples were washed and fixed for immunofluorescence assays. The observation and capture of images was performed with a LSM-50 Confocal Scanning Microscope (Carl Zeiss, Germany).
Immunofluorescence assay
RAGE and S100B were detected in HUVEC’s exposed to inflammatory stimuli. The observation and analysis were performed using immunofluorescence staining with specific antibodies (Abcam Co., Cambridge, Massachusetts, USA). For the in vitro assays, HUVEC’s were washed three times with PBS at pH 7.2, fixed in 4% paraformaldehyde (PFA) for 15 minutes, and washed three times with PBS. These preparations were incubated in 0.6 M glycine for 20 minutes, to remove the autofluorescence of aldehydes and washed three times with PBA (PBS albumin, 0.5%). Samples were then blocked for 20 minutes with PBA 1% to eliminate nonspecific reactions of the antibodies, and subsequently washed three times with PBA 0.5%. Immunoreactions were carried out diluting the mouse primary antibody specific for S100B 1:100 (SH-B1, ab11178, Biotech, Cambridge, UK), or the polyclonal rabbit antibody for RAGE (ab3611, Biotech) in PBA 1%, and cells or tissue slices were incubated for 1 hour at 4 °C. They were subsequently washed three times with PBA 1 %. The secondary antibodies (Alexa 647 for S100B and Alexa-488 for RAGE, Life Technologies) were diluted 1:400 and incubated with the samples for 1 h; afterwards, the preparations were washed with PBA 1%. Nuclei were stained blue with DAPI (Vectashield, Vector Laboratories, Inc., Burlingame, California, USA) and visualized using a fluorescent microscope (Floid Cell Imaging Station, Life Technologies).
Image processing
In order to estimate the co-localization of RAGE and S100B in cells, the merge area from the fluorescent labeling was analyzed by Otsu’s method for segmentation of the region of interest [28]. A value of 50 is used as a threshold for the red and green color image components. Contours are found applying a Laplacian operator of 3 x 3 pixels to the binary image after segmentation. The area is represented by the number of pixels different to the background in the binary image. The average of the intensity is computed in the region of interest. Z-stack processing was also employed for depth improvement of images containing HUVEC’s. This method is commonly used to improve the analysis of three-dimensional images, hence allowing more detailed recognition of cellular structures and better localization of proteins.
Statistical analysis
All experiments were carried out in duplicate. Mean ± standard error medium (S.E.M.) were determined for all parameters. Statistical significance among groups was determined by Student’s t-test or one-way analysis of variance followed by Bonferroni’s post-hoc analysis. Values of P<0.05 were considered as statistically significant.
Results
LPS induced an early and enhanced co-expression of RAGE and S100B in HUVEC’s after 3 h of exposure
Fig. 1 shows the co-localization of RAGE and S100B in HUVEC’s exposed for 3 h to pro-inflammatory stimuli induced by LPS or TNF-α (as a positive control). Cells were labeled for immunofluorescence in blue (DAPI nuclei), green (RAGE) and red (S100B). The first line corresponds to the sequence of untreated (control) cells, where moderate co-localization areas of both proteins can be observed in the merge (Fig. 1 A). In addition, in these cells, the segmentation method revealed the specific points of co-localization evidenced by a labeling area (16230 pixels per field). Quantification of these co-localization areas was achieved also with the segmentation method (Fig. 1 A-B). The second sequence of images (second line) depicts the effect of LPS on RAGE and S100B co-localization; this treatment produced an increased number of labeled cells for both proteins when co-localized in the merge, as compared to untreated cells. Concomitantly, the RAGE and S100B co-localization areas (segmentation) in LPS-treated cells were enhanced when compared with control cells (186 % above the control). In the third line, the sequence of images shows that the exposure of cells to TNF-α produced a prominent RAGE and S100B co-localization (in the merge), as well as an increased labeling area (334 % above the control).
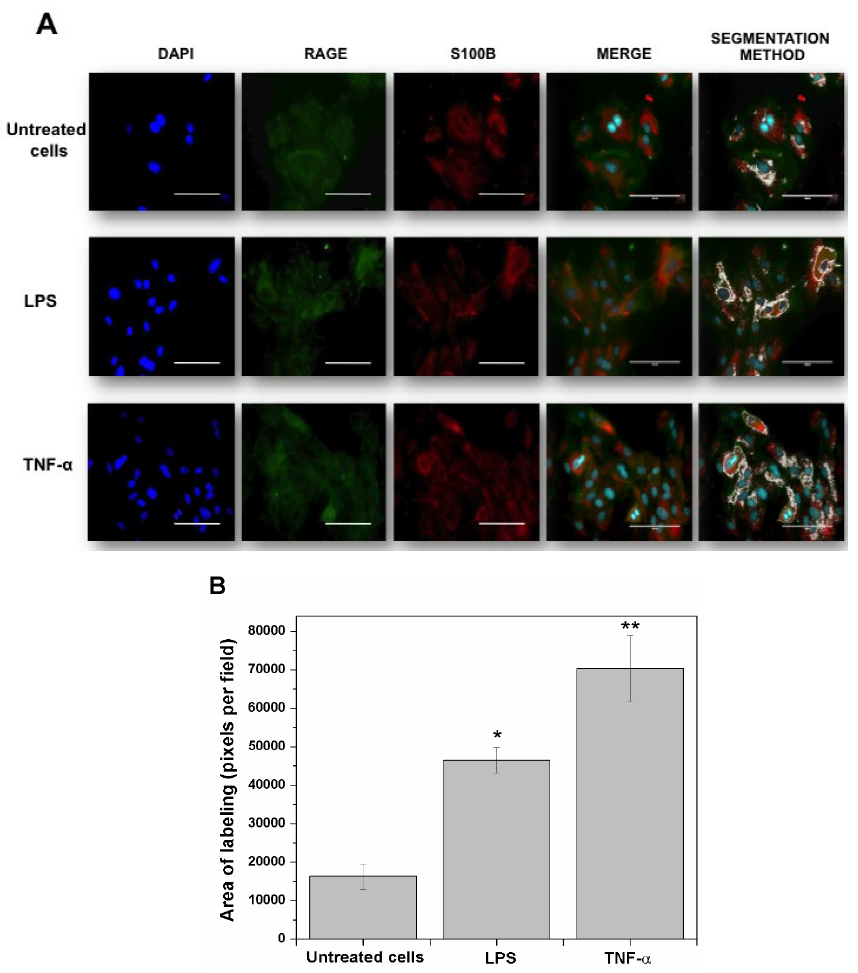
Fig. 1 RAGE and S100B co-localization in human umbilical vein endothelial cells (HUVEC’s) obtained from umbilical cord. Cells were challenged with acute inflammatory stimuli (TNF-α [5 ng/ml] or LPS [10 ng/ml]). RAGE and S100B were detected early by immunofluorescence after exposure to pro-inflammatory agents for 3 h. In A, the localization of RAGE (green column) and S100B (red column) are shown. Nuclei were stained in blue (DAPI). In the fourth and fifth columns, the images show the co-localization (merge) and image analysis (segmentation algorithm) of both proteins, respectively. The horizontal bars indicate a length of 100 µm in all panels. In B, the graphic representation of the merge area (as percent) in untreated (Control) and TNF-α- or LPS-treated cells is shown. Bars represent quantification of two images per treatment. One-way ANOVA followed by Bonferroni’s test; *p<0.05, **p<0.01, different from untreated cells.
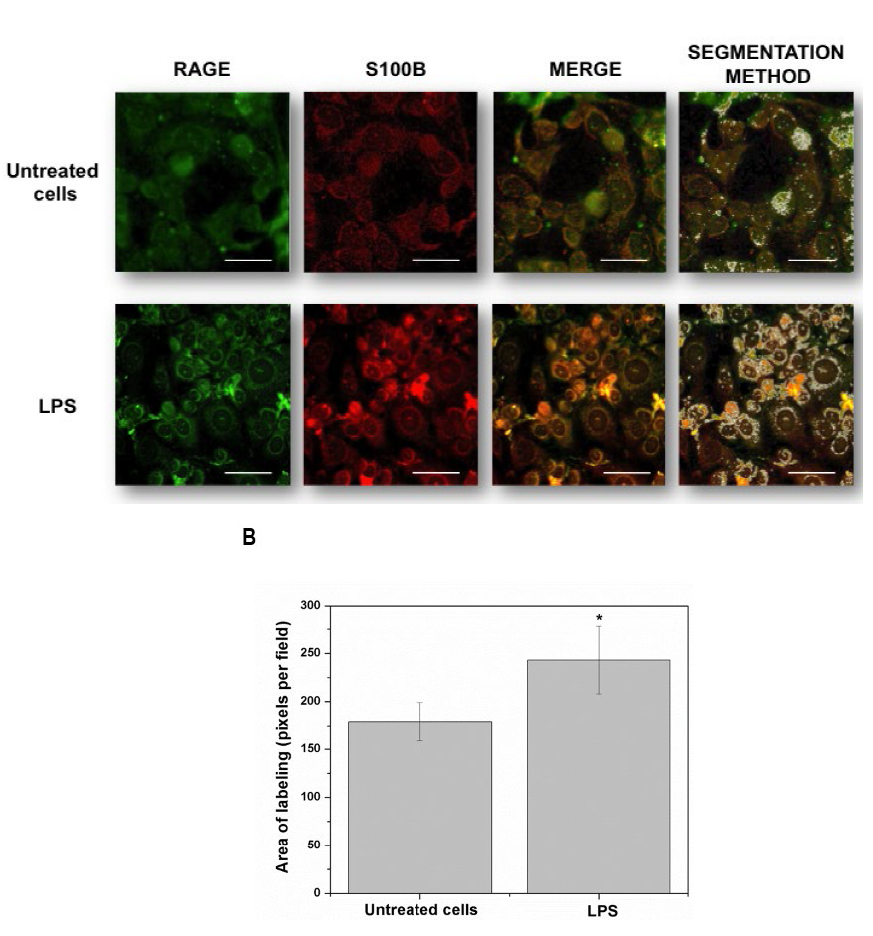
Fig. 2 Detailed images (20 µm bar scale) showing untreated cells (upper line) and LPS-treated HUVEC’s (lower line). In A, Images correspond to three-dimensional reconstructions using the Z-stack to display the expression of RAGE (first column in green), S100B (second column in red) and the merge (third column). RAGE was constructed with 18 photos, whereas S100B was built up with 12 photos. The fourth column depicts the segmentation in the co-localization images. In B, the graphic representation of the merge area (as percent) in untreated (Control) and LPS-treated cells is shown. Bars represent quantification of two images per treatment. Student’s t-test; *p<0.05, different from untreated cells.
Discussion
Endothelial dysfunction and inflammatory cell adhesion have been regarded as early and important factors in various pathologies. In fact, inflammation is a major event for both, the healing and the damaging processes. In turn, LPS is a pro-inflammatory molecule involved in the pathogenesis of inflammatory disorders. While chronic inflammation is more commonly associated with pathological stages, the characterization of acute inflammation is of major relevance since this event establishes the borders between healing and injury. How cells and tissues respond to acute inflammation is the clue to anticipate whether they will survive or initiate deadly cascades, leading further to tissue damage. Therefore, in this study we considered the early effects (3 h exposure) of LPS on HUVEC’s in order to complement previous reports describing its actions on cell viability at different times (1 and 24 h) [29]. Further support for an early role of LPS and TNF-α as inflammatory promoters for HUVEC’s can be found in related reports [30,31]. The response of endothelial cells in vitro was sensed after LPS stimulation upon the assumption that this pro-inflammatory molecule binds to Toll-like membrane receptors and RAGE, thereby inducing changes in their phenotype at short times [32].
Here, we demonstrated an early co-localization of RAGE and S100B proteins in HUVEC’s subjected to inflammatory stimuli, thus suggesting that this interaction might eventually be responsible for inflammatory events through signaling cascades involving factors like NFκB [33]. In this regard, several studies have suggested that RAGE activation during inflammatory processes involving the S100B protein, is taking place [34]. Indeed, the elicited inflammatory cascades induced by RAGE probably perpetuate the organ dysfunction in inflammatory processes [6]. The interaction of these two proteins for triggering acute and/or chronic inflammation and its biological role has already been characterized [7,35,36]; yet, a still unresolved issue is the minimal size of the S100B-RAGE complex that can promote signaling, bringing together the cytoplasmic tails of the RAGE dimers.
Despite it being currently assumed that different ligands can bind to RAGE, directly or through other molecules (i.e., forming bridges with AGEs) [37], all possible interactions between these molecules or their effects on cells, have not been explored yet. RAGE activation by S100B is a major pro-inflammatory pathway occurring in acute and chronic inflammatory diseases [23,38]. Some members of the S100 protein family, such as S100B, are known to form active tetramers, hexamers or larger oligomers [7,39].
The co-localized areas for RAGE and S100B could suggest either an increase in the expression of these proteins, or their mere re-location with additional interactions. It is therefore fair to mention that our results are focused exclusively on S100B from endothelial cells, where this protein can act as a chemokine that is subsequently released to the surrounding environment. Based on these conditions, the characterization of the interactions between RAGE and S100B and their effects are of major relevance, since they can serve for the design of more specific in vivo models of disorders involving inflammatory components.
Finally, the occurrence of the RAGE-S100B interaction in HUVEC’s establishes the basis for the understanding of the inflammatory events taking place during the gestational and/or birth processes under normal or pathological conditions, though such interaction has also been described in other biological preparations, including different tumor cells, neurons, astrocytes, microglia, etc. This is relevant as RAGE has been shown to interact with several other ligands, including diverse members of the S100 protein family, thus participating in disorders like cancer, cardiovascular disease, CNS disorders, etc. [40,41]. Therefore, the many biological implications that the RAGE-S100B interaction represent for the compromise of physiological homeostasis deserves deeper and more detailed investigation in different biological scenarios











 nueva página del texto (beta)
nueva página del texto (beta)


