Celebrating Prof. Víctor M. Loyola Vargas career.
In our work, we preserve the vigour and passion with which you taught us to live in the laboratory.
Introduction
The plant family Cactaceae comprises around 1400 succulent and non-succulent species distributed throughout the entire American continent [1]. In Mexico, more than 150 species of cacti are used by indigenous people, and at least 50 of them are cultivated [2], some species such as Opuntia ficus-indica, Hylocereus undatus, and Acanthocereus tetragonus, stand out for their economic importance as producers of food, forage, and dyes [3-6]. Recently, there has been an increasing interest in searching for metabolites in some cactus species in order to apply that knowledge in the industry. The analysis of methanolic extracts of crude and cooked stems of A. tetragonus -using ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry indicated the presence of carboxylic acids such as threonic, citric, and malic acids, phenolic acids, and glycosylated flavonoids (luteolin-O-rutinoside). The proportions of citric acid found within these compounds are noteworthy because this organic acid is highly demanded by the food and beverages, pharmaceutical, and personal care industries [5]. HPLC analysis results in three Opuntia species (O. ficus-indica, O. undulata, and O. stricta) detected ascorbic acid, flavonoids (quercetin, isorhamnetin, myricetin, kaempferol, and luteolin), betalains, taurine, total carotenoids, and total phenolics. Another study showed the presence of flavonoids in four cultivars of O. ficus-indica [6]. The family Cactaceae has been little explored at the genomic and transcriptomic level despite the proven commercial importance of some species in the family. There are reports of transcriptomic studies made in Melocactus glaucescens, Mammillaria bombycina, and Lophophora williamsii. In these studies, the objectives were to discover metabolic changes during in vitro shoot organogenesis induction [7], study responses to abiotic stress [8], and search for genes putatively involved in mescaline biosynthesis [9]. Bioinformatic analysis is a tool that could allow us to predict the presence of protein-coding genes -including enzymes- and to know how these genes are functionally related in the different metabolic pathways of an organism. Using this strategy, we may be able to identify genes putatively involved in biosynthetic pathways of interest to produce metabolites that are important in the industry and, in the future, to improve the production of compounds of commercial interest through genetic engineering strategies [10,11].
Because of the local economic and ecological importance of the family Cactaceae in Mexico and the American continent, its species could be excellent models for searching for genes encoding enzymes involved in secondary metabolism. There needs to be more exploration of genes involved in secondary metabolism in cacti. Public databases such as that of the National Center for Biotechnology Information (NCBI) contain little information about genes annotated and identified in members of the family Cactaceae. However, the genomes of Carnegiea gigantea, Cereus fernambucensis, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, Selenicereus undatus, and Stenocereus thurberi have been sequenced [12] and can be explored to identify genes putatively involved in the secondary metabolism of cacti. Cacti have ecological importance due to their ability to survive in extreme environments, high temperatures, geographical areas with little rainfall, and on arid soils [13]. CAM metabolism and C3 or C4 carbon fixation are known to be present in members of the cactus family [1]. The seven cacti species that we analyzed in this work have received recent attention aimed at obtaining bioactive compounds from them [14,15]. Cereus jamacaru contains a large diversity of biological compounds, including alkaloids, steroids, triterpenes, glycosides, oils, and waxes used by the pharmaceutical industry [14]. Lophocereus schottii is used in Mexico for traditional medicine cancer treatment. The polar fraction of its ethanolic extract contains flavonoids, alkaloids, terpenoids, and sterols with a proven cytotoxic effect on L5178Y cells and splenocytes stimulated with mitogen concanavalin A (ConA) [15]. In Central America and Mexico, the fruits of Hylocereus undatus, H. costaricensis, H. polyrhizus, and Selenicereus megalanthus -dragon fruit or pitahayas in Mexico- are traded and consumed by people [16]. Studies aimed at identifying bioactive compounds in this genus have indicated that the ethanolic crude extract of the fruit of Selenicereus undatus helps to normalize glucose homeostasis in mice, suggesting that the active compounds may be helpful in the management of diabetes mellitus [17]. A study comparing the metabolomic profiles of Hylocereus polyrhizus and Hylocereus undatus emphasized the value as raw matter in the pharmaceutical and food industries of the bio-residual skins of the pitahaya due to the presence in them of active compounds (L-tyrosine, L-valine, DL-norvaline, tryptophan, γ-linolenic acid, and isorhamnetin-3-O-neohesperidoside) [16]. Alkaloids are one of the most commercially successful groups of molecules because of their antioxidant, antibacterial, antiparasitic, insecticidal, anticorrosive, and antiplasmodial activities [18,19]. Regarding genomic studies of cactus aimed at identifying genes encoding enzymes related to metabolic pathways, only two reports are available, one in which the H. undatus genome was annotated and compared with that of Carnegiea gigantea, finding that nearly 29,000 protein-coding genes were similar between both genomes [12], and another one that identified genes coding for enzymes involved in the betacyanin biosynthetic pathway -which in the future might be used in genetic engineering to improve fruit colour in species with white (Hylocereus undatus) or yellow (Selenicereus megalanthus) pulp [9]- and identified the genes involved in mescaline biosynthesis using de novo sequencing and transcriptome analysis in Lophophora williamsii.
Our review of the literature revealed that cacti genomes have not been extensively explored to identify genes involved in secondary metabolism at the genetic level. Therefore, in this research we annotated genes putatively encoding enzymes involved in secondary metabolism in Carnegiea gigantea, Cereus fernambucensis, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, Selenicereus undatus, and Stenocereus thurberi.
Experimental
Materials and methods
Phylogenetic analysis of the family Cactaceae
To know the evolutionary relationships between members of the family Cactaceae, in this study, we analyzed 25 partial amino acids (aa) sequences of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39; RuBisco). The evolutionary history was inferred using the Minimal Evolution (ME) method [20]. Evolutionary analyses were performed on MEGA11 [21]. The percentage of replicate trees in which associated taxa clustered in the bootstrap test (1000 replicates) is shown below the branches [22]. Evolutionary distances were calculated using the Poisson correction method [23] and expressed in units of the number of aa substitutions per site. The ME tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1 [24]. The neighbor-joining algorithm [25] was used to generate the initial tree. All ambiguous positions were removed for each pair of sequences (pairwise removal option). There was a total of 460 positions in the final data set.
Assembly and annotation of the genome of Cereus fernambucensis
The genome of Cereus fernambucensis was obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/data-hub/genome/?taxon=308225). The set of sequences was made with DRAM and the annotations, with the DomainAnnotation v1.0 program that identifies protein domains from domain libraries. For this, the comparison of the C. fernambucensis genome was made with all the domain libraries of the platform of the Knowledge base of the Department of Biology of Energy Systems (KBase). The domain libraries in the KBase include: 1. the NCBI Conserved Domain Database (CDD), COGs (Clusters of Orthologous Groups). 2. NCBI CDD models from NCBI Conserved Domain Database version 3.16 (CDD) [26]. These libraries only contain domains curated by NCBI (including "sd" structural motif models). 3. the SMART (Simple Modular Architecture Research Tool) version 6.0 [27], from CDD. 4. PRK (Protein Clusters version 6.0, from CDD. 4. Pfam version 31.0 hidden Markov models (HMM) [28]. 5. TIGRFAMs version 15.0 HMM [29,30] from the J. Craig Venter Institute. 6. NCBIfam version 1.1 HMM. For the first four libraries mentioned above (COG, CDD, SMART, and PRK), KBase runs RPS-BLAST version 2.2.31 [31] of the BLAST+ package at NCBI [32], identifying all domain accesses with an E value of 10-4 or better. For the three HMM libraries (Pfam, TIGRFAMs, and NCBIfam), KBase runs HMMER version 3.1b2, identifying all domain results at least as significant as the family-specific confidence bound identified by the curators of each model.
Identification of protein domains involved in the secondary metabolism of members of the family Cactaceae
From the annotation of the genome of C. fernambucensis (WGS accession JALPLW01 and GenBank GCA_024363205.1), the enzymes involved in plant secondary metabolism were identified according to their biosynthetic origin in seven functional groups: alkaloids and nitrogenous compounds, flavonoids, terpenes, phenolic compounds, shikimate pathway, ABC-type transporters, and proteins involved in defense against plant pathogens. In order to learn the evolutionary relationships of different enzymes for each functional group, the genomes of C. fernambucensis, Selenicereus undatus, Stenocereus thurberi, Lophocereus schottii, Pachycereus pringlei, Pereskia humboldtii, and Carnegiea gigantea, and other accessions of eudicots and flowering plants from the NCBI database, were compared with the enzymes plant PDR-type ABC transporter family protein (PEN3) from Arabidopsis thaliana, aspartic proteinase-like protein, flavanone 3-hydroxylase (F3H), hydroxycinnamoyl-CoA shikimate quinate hydroxycinnamoyl transferase (HCT), serine threonine-protein phosphatase, and sterol methyltransferase (SMT1). Dendrograms were constructed in MEGA11 with the Minimal Evolution method using the aa sequences of the structural domains of the proteins of interest. For each analysis, the optimal tree is shown in which the percentage of replicate trees in which associated taxa clustered in the bootstrap test (1000 replicates) is shown next to evolutionary branches.
Tertiary structure of enzymes in members of the Cactaceae family
The analyses of the tertiary structure of flavanone 3-hydroxylase (F3H) were obtained from the enzyme's sequences of nucleotides and aa in SWISS-MODEL (https://swissmodel.expasy.org/) [33], considering the sequence identity values (%), Global Model Quality Estimate (GMQE), and Qualitative Model Energy Analysis (QMEAN). Ten plant accessions (Gossypium arboreum (XP_017638014.1), Vernicia fordii (ARV78456.1), Hibiscus sabdariffa (ALB35017.1), Durio zibethinus (XP_022741107.1), Althaea officinalis (UOI87842.1), Dimocarpus longan (ABO48521.1), Arachis hypogaea (XP_025694592.1), Boehmeria nivea (QBC98316.1), Ziziphus jujuba (XP_015889639.1), and Litchi chinensis (ADO95201.1) were included for the comparative analysis of the 3D conformation and the effect of aa substitutions in the 2-oxoglutarate iron-dependent domain. The three-dimensional modeling was created in the Unipro UGENE v. 45.0 platform from the consensus sequence of the regions obtained in the analysis of the genomes of accessions identified as members of the family Cactaceae, and comparison with the reference model described in [34] for anthocyanidin synthase 1gp4.1A in Arabidopsis thaliana.
Results and discussion
Phylogenetic relationships in the family Cactaceae
Among the genera in the family Cactaceae that we analyzed in this work, the genus Cereus had the highest number of accepted and registered taxa with 187 species, 8 subspecies, 12 varieties, and 10 hybrids, followed by Selenicereus (36 species and 6 subspecies), Stenocereus (25 species and 2 subspecies), Pereskia (19 species and 4 subspecies), and, with fewer recorded species, Lophocereus, Pachycereus, and Carnegiea with 7, 4 and 2 species, respectively [35].
In the NCBI database, seven Cacteaceae genomes are available: JALPLW01 (Cereus fernambucensis), JACYFF01 (Selenicereus undatus), NCQT01 (Stenocereus thurberi), NCQV01 (Lophocereus schottii-), NCQS01 (Pachycereus pringlei), NCQU01 (Pereskia humboldtii), and NCQR01 (Carnegiea gigantea). To gain some knowledge on the evolutionary relationships of these species, a phylogenetic analysis was performed using a conserved domain of the small subunit of the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisco) involved in carbon fixation to an organic form (glucose) during the Calvin cycle and in photorespiration. RuBisco catalyzes the initial steps of photosynthetic carbon reduction and photorespiratory carbon oxidation [36,37]. Land plants have an hexadecameric type I RuBisco with an approximate molecular mass of 550 kDa, composed of eight large (L;55 kDa) and eight small (S;15 kDa) subunits (L8S8). In A. thaliana, four distinct small-subunit isoforms of RuBisco are known (RbcS1A, RbcS1B, RbcS2B and RbcS3B) [38].
Our analysis of the phylogenetic relationships of 25 species in the family Cactaceae indicated the formation of two clades, the first one grouped Pereskia humboldtii (NCQU01.1), Pereskia aculeata (YP010365471.1), Pachycereus pringle (NCQS01079793.1), Pereskiopsis diguetti (AFB70629.1), Maihuenia poeppigii (AFB70626.1) and Opuntia sp. (UEK25784.1, QJT42891.1), and the second one included Cereus fernambucensis (JALPLW010162134.1), Selenicereus undatus (YP010023660.1), Lophocereus schottii (YP009590644.1), Carnegiea gigantea (AKR06847.1), and Stenocereus thurberi (UXN84199.1), which grouped together with species of Ferocactus (QTI91393.1, QTI91309.1), Mammillaria (QFG71193.1, QFG71129.1), and Rhipsalis (YP010015700, YP010166122.1). Within this group, we observed that Ferocactus setispinus and Ferocactus latispinus are relatively young species compared to the other accessions in this clade (Fig. 1). The genus Pereskia (Cactaceae) comprises 19 species. Recent phylogenetic work has shown that the species in this genus group into two separate clades, which are basal and paraphyletic with respect to the rest of the cacti [39], therefore it is possible that members of Pereskia -Pereskia humboldtii (NCQU01.1) and Pereskia aculeata (YP010365471.1)- form an independent clade of more closely related cacti. It is interesting to observe that, apparently, Pereskia is more closely related to species of Opuntia (O. sulphurea and O. quimilo). In the other clade it was possible to observe that most of the genomes analyzed in this work (Cereus fernambucensis (JALPLW010162134.1), Selenicereus undatus (YP010023660.1), Lophocereus schottii (YP009590644.1), Carnegiea gigantea (AKR06847.1), Stenocereus thurberi (UXN84199.1), present a closer grouping (Fig. 1). There are few investigations in which the phylogenetic relationship of cacti are based on specific regions of constitutive genes (like RuBisco), which are genes that have demonstrated conservation at the genomic and structural levels, and that allow us to observe groupings associated to phylogeny in various plant genera with C3 and C4 carbon fixation [40, 41]. A study where the phylogeny of the genus Leptocereus (Cactaceae) was reconstructed using chloroplastid markers (trnL-F, trnQ-rps16, psbA-trnH, petL-psbE, and rpl16), indicated that the genus Leptocereus has a paraphyletic origin, and proved to be phylogenetically distant from some members belonging to the genus Selenicereus (S. grandiflorus and S. calcaratus, therefore demonstrating that there is no close resolution among these members of this genus [42].
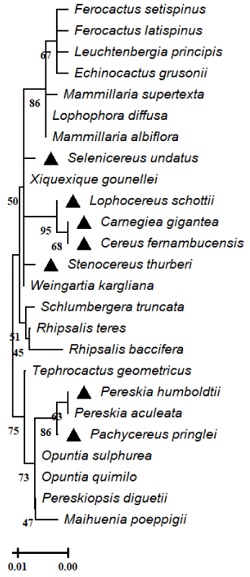
Fig. 1 Phylogenetic analysis of the RuBisco partial small subunit in the family Cactaceae. The evolutionary relationships were carried out using the MEGA program with the method of minimum evolution and with a bootstrap of 1000 repetitions. Partial sequences of 460 amino acid residues from the accessions Carnegiea gigantea (AKR06847.1), Lophocereus schottii (YP 009590644.1), Stenocereus thurberi (UXN84199.1), Selenicereus undatus (YP 010023660.1), Rhipsalis teres (YP 010166122.1), Mammillaria albiflora (QFG71193.1), Schlumbergera truncata (P25835.1), Lophophora diffusa (YP 010411775.1), Tephrocactus geometricus (YP 010411685.1), Echinocactus grusonii (QTI91172.1), Mammillaria supertexta (QFG71129.1), Opuntia sulphurea (UEK25784.1), Leuchtenbergia principis (QTI91241.1), Opuntia quimilo (QJT42891.1), Ferocactus setispinus (QTI91393.1), Rhipsalis baccifera (YP 010015700.1), Ferocactus latispinus (QTI91309.1), Weingartia kargliana (AFB70631.1), Pereskia aculeata (NCQU01.1), Pereskiopsis diguetii (AFB70629.1), Maihuenia poeppigii (AFB70626.1), Pereskia aculeata (AAL35687.1), Xiquexique gounellei (QKN21858.1), Cereus fernambucensis (JALPLW010162134.1) and Pachycereus pringlei (NCQS01079793.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
One of the disadvantages when carrying out this type of molecular analysis is the scant information deposited in public databases, as well as the identification, amplification, and sequencing of specific fragments of the genome of species belonging to the genera of the family Cactaceae. However, derived from the advances in molecular techniques and sequencing platforms, it should be important to highlight the importance of directing studies of phylogenetic reconstructions in cacti through molecular markers. In this study we demonstrate that the use of RuBisco as a molecular marker for cacti phylogeny could be applied in the future, including a greater number of representative species of each genus in the Cactaceae family.
Enzymes involved in the biosynthesis of secondary metabolites in the genome of C. fernambucensis
The genome of C. fernambucensis with the accession WGS JALPLW01 was assembled with N50 the KBase 489.3 Mb platform with 177,285 contigs and 298,226 CDS numbers. It scanned 298,226 protein sequences for 5892,916 characteristic kmers and found 19,252 characteristic kmers and was able to predict 373 enzyme functions for 1,773 protein sequences. This result indicates that for this set of protein sequences, 73 % of the enzymatic functions of the primary metabolism of plants were detected (See Table 1, Supplementary Fig. 1). The reports of enzymes involved in secondary metabolism pointed to the identification of domains cd07816, smart01037, cd10810 of the family of cytoplasmic proteins related to plant pathogenesis (PR-10) -very widespread among dicotyledonous plants, norcoclaurine synthases (NCS), cytokinin-binding proteins (CSBPs), major latex proteins (MLPs), and maturation-related proteins. We also identified CDS of aldehyde reductase, flavonoid reductase of the extended SDR type and related proteins (cd05193, cd08958), related flavonoid reductases acting in the NADP-dependent reduction of flavonoids, ketone-containing secondary plant metabolites, terpene cyclases, isoprenoid biosynthesis enzymes (cd00868) of the class 1 superfamily, and a diverse group of monomeric plant terpene cyclases (Tspa-Tspf) converting the acyclic isoprenoid diphosphates, geranyl diphosphate (GPP), farnesyl diphosphate (FPP), or geranylgeranyl diphosphate (GGPP) into cyclic monoterpenes, diterpenes, or sesquiterpenes (cd00684). We also found CDS of enzymes involved in the biosynthesis of phenolic compounds, such as laccase (cd13897, cd13849) -a blue multicopper oxidase (MCO) which catalyzes the oxidation of a variety aromatic phenolic and inorganic substances coupled to the reduction of molecular oxygen to water, and of the shikimic acid pathway -of great relevance in the biosynthesis of the three aromatic amino acids phenylalanine, tyrosine and tryptophan, as well as a wide range of secondary metabolites (cd00502, COG0169, COG0703, cd01065, PF01488.19). Interestingly, a large number of ABC transport system domains were found (cd03264, cd03235, cd03246, cd03297, cd03300, COG1127, cd03369, cd02079, cd00994, cd02076, cd01431, cd03227, cd06340, cd06349). ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds like sugars, ions, peptides, and more complex organic molecules. This family includes transporters involved in the uptake of various metallic cations such as iron, manganese, and zinc.
Table 1 Enzymatic secondary metabolism routes found of Cereus fernambucensis.
| Secondary metabolite route | Domain | Conserved Protein Domain Family | Description |
| Alkaloids and nitrogenous compounds | cd10810 | Pathogenesis-related protein Bet v I family. | Pathogenesis-related proteins PR-10: These proteins were identified as major tree pollen allergens in birch and related species (hazel, alder), as plant food allergens expressed in high levels in fruits, vegetables and seeds (apple, celery, hazelnut), and as pathogenesis-related proteins whose expression is induced by pathogen infection, wounding, or abiotic stress. Hyp-1 (Q8H1L1), an enzyme involved in the synthesis of the bioactive naphthodianthrone hypericin in St. John's wort ( Hypericum perforatum ) also belongs to this family. Most of these proteins were found in dicotyledonous plants. In addition, related sequences were identified in monocots and conifers. Cytokinin-specific binding proteins: These legume proteins bind cytokinin plant hormones [(PUBMED:9874249)]. (S)-Norcoclaurine synthases are enzymes catalyzing the condensation of dopamine and 4-hydroxyphenylacetaldehyde to (S)-norcoclaurine, the first committed step in the biosynthesis of benzylisoquinoline alkaloids such as morphine (PUBMED:15447655). Major latex proteins and ripening-related proteins are proteins of unknown biological function that were first discovered in the latex of opium poppy ( Papaver somniferum ) and later found to be upregulated during ripening of fruits such as strawberry and cucumber [ (PUBMED:15447655)]. The occurrence of Bet v 1-related proteins is confined to seed plants with the exception of a cytokinin-binding protein from the moss Physcomitrella patens (Q9AXI3). |
| cd07816 | Ligand-binding bet_v_1 domain of major pollen allergen of white birch ( Betula verrucosa ), Bet v 1, and related proteins. | This family includes the ligand binding domain of Bet v 1 (the major pollen allergen of white birch, Betula verrucosa ) and related proteins. In addition to birch Bet v 1, this family includes other plant intracellular pathogenesis-related class 10 (PR-10) proteins, norcoclaurine synthases (NCSs), cytokinin binding proteins (CSBPs), major latex proteins (MLPs), and ripening-related proteins. | |
| Flavonoids | cd05193 | Aldehyde reductase, flavonoid reductase, and related proteins, extended (e) the serine-aspartate repeat protein family (SDRs). | This subgroup contains aldehyde reductase and flavonoid reductase of the extended SDR-type and related proteins. The related flavonoid reductases act in the NADP-dependent reduction of flavonoids, ketone-containing plant secondary metabolites. Extended SDRs are distinct from classical SDRs. |
| cd08958 | Flavonoid reductase (FR), extended (e) SDRs. | This subgroup contains FRs of the extended SDR-type and related proteins. These FRs act in the NADP-dependent reduction of flavonoids, ketone-containing plant secondary metabolites; they have the characteristic active site triad of the SDRs (though not the upstream active site Asn) and a NADP-binding motif that is very similar to the typical extended SDR motif. | |
| Terpenes | cd00684 | Plant terpene cyclases, class 1. | This CD includes a diverse group of monomeric plant terpene cyclases (Tspa-Tspf) that convert the acyclic isoprenoid diphosphates, geranyl diphosphate (GPP), farnesyl diphosphate (FPP), or geranylgeranyl diphosphate (GGPP) into cyclic monoterpenes, diterpenes, or sesquiterpenes, respectively; a few form acyclic species. |
| cd00868 | Terpene cyclases, class 1. | Terpene cyclases, class 1 (C1) of the class 1 family of isoprenoid biosynthesis enzymes, which share the 'isoprenoid synthase fold' and convert linear, all-trans, isoprenoids, geranyl (C10)-, farnesyl (C15)-, or geranylgeranyl (C20)-diphosphate into numerous cyclic forms of monoterpenes, diterpenes, and sesquiterpenes. Also included in this CD are the cis-trans terpene cyclases such as trichodiene synthase. | |
| cd02892 | Squalene cyclase (SQCY) domain subgroup 1. | Found in class II terpene cyclases that have an alpha 6 - alpha 6 barrel fold. Squalene cyclase (SQCY) and 2,3-oxidosqualene cyclase (OSQCY) are integral membrane proteins that catalyze a cationic cyclization cascade converting linear triterpenes to fused ring compounds. | |
| cd00385 | Isoprenoid biosynthesis enzymes, class 1. | Superfamily of trans-isoprenyl diphosphate synthases (IPPS) and class I terpene cyclases which either synthesis geranyl/farnesyl diphosphates (GPP/FPP) or longer chained products from isoprene precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), or use geranyl (C10)-, farnesyl (C15)-, or geranylgeranyl (C20)-diphosphate as substrate. These enzymes produce a myriad of precursors for such end products as steroids, cholesterol, sesquiterpenes, heme, carotenoids, retinoids, and diterpenes. | |
| cd00867 | Trans-Isoprenyl Diphosphate Synthases. | ||
| cd00685 | Trans-Isoprenyl Diphosphate Synthases, head-to-tail. | These trans-isoprenyl diphosphate synthases (trans_IPPS) catalyze head-to-tail (HT) (1'-4) condensation reactions. This CD includes all-trans (E)-isoprenyl diphosphate synthases which synthesize various chain lengths (C10, C15, C20, C25, C30, C35, C40, C45, and C50) linear isoprenyl diphosphates from precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Farnesyl diphosphate synthases produce the precursors of steroids, cholesterol, sesquiterpenes, farnsylated proteins, heme, and vitamin K12; and geranylgeranyl diphosphate and longer chain synthases produce the precursors of carotenoids, retinoids, diterpenes, geranylgeranylated chlorophylls, ubiquinone, and archaeal ether linked lipids. | |
| Phenolic compounds | cd13897 | The third cupredoxin domain of the plant laccases. | Laccase is a blue multicopper oxidase (MCO) which catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. Laccase has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. |
| cd13849 | The first cupredoxin domain of plant laccases. | ||
| Shikimate pathway | cd00502 | Type I 3-dehydroquinase, (3-dehydroquinate dehydratase or DHQase). | Dehydroquinase is the third enzyme in the shikimate pathway, which is involved in the biosynthesis of aromatic amino acids. Type I DHQase exists as a homodimer. |
| COG0169 | Shikimate 5-dehydrogenase. | Amino acid transport and metabolism. | |
| COG0703 | Shikimate kinase. | ||
| cd05213 | NADP-binding domain of glutamyl-tRNA reductase. | Glutamyl-tRNA reductase catalyzes the conversion of glutamyl-tRNA to glutamate-1-semialdehyde, initiating the synthesis of tetrapyrrole. Whereas tRNAs are generally associated with peptide bond formation in protein translation, here the tRNA activates glutamate in the initiation of tetrapyrrole biosynthesis. | |
| cd01065 | NAD(P) binding domain of Shikimate dehydrogenase. | Shikimate dehydrogenase (DH) is an amino acid DH family member. Shikimate pathway links metabolism of carbohydrates to de novo biosynthesis of aromatic amino acids, quinones and folate. | |
| PF01488.19 | Shikimate / quinate 5-dehydrogenase. | This family contains both shikimate and quinate dehydrogenases. Shikimate 5-dehydrogenase catalyses the conversion of shikimate to 5-dehydroshikimate. This reaction is part of the shikimate pathway which is involved in the biosynthesis of aromatic amino acids. Quinate 5-dehydrogenase catalyses the conversion of quinate to 5-dehydroquinate. This reaction is part of the quinate pathway where quinic acid is exploited as a source of carbon. Both the shikimate and quinate pathways share two common pathway metabolites 3-dehydroquinate and dehydroshikimate. | |
| cd05312 | NAD(P) binding domain of malic enzyme (ME), subgroup 1. | Malic enzyme (ME), a member of the amino acid dehydrogenase (DH)-like domain family, catalyzes the oxidative decarboxylation of L-malate to pyruvate in the presence of cations (typically Mg++ or Mn++) with the concomitant reduction of cofactor NAD+ or NADP+. ME has been found in all organisms and plays important roles in diverse metabolic pathways such as photosynthesis and lipogenesis. This enzyme generally forms homotetramers. | |
| cd01079 | NAD binding domain of methylene-tetrahydrofolate dehydrogenase. | The NAD-binding domain of methylene-tetrahydrofolate dehydrogenase (m-THF DH). M-THF is a versatile carrier of activated one-carbon units. | |
| cd05212 | NAD(P) binding domain of methylene-tetrahydrofolate dehydrogenase and methylene-tetrahydrofolate dehydrogenase/cyclohydrolase. | ||
| Plant transporters | cd03264 | ABC-type multidrug transport system, ATPase component. | The biological function of this family is not well characterized, but display ABC domains similar to members of ABCA subfamily. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides, and more complex organic molecules. |
| cd03235 | ATP-binding cassette domain of the metal-type transporters. | This family includes transporters involved in the uptake of various metallic cations such as iron, manganese, and zinc. The ATPases of this group of transporters are very similar to members of iron-siderophore uptake family suggesting that they share a common ancestor. | |
| cd03246 | ATP-binding cassette domain of PrtD, subfamily C. | This family represents the ABC component of the protease secretion system PrtD, a 60-kDa integral membrane protein sharing 37% identity with HlyB, the ABC component of the alpha-hemolysin secretion pathway, in the C-terminal domain. They export degradative enzymes by using a type I protein secretion system and lack an N-terminal signal peptide but contain a C-terminal secretion signal. | |
| cd03297 | ATP-binding cassette domain of the molybdenum transport system. | ModC is an ABC-type transporter and the ATPase component of a molybdate transport system that also includes the periplasmic binding protein ModA and the membrane protein ModB. ABC transporters are a large family of proteins involved in the transport of a wide variety of different compounds, like sugars, ions, peptides and more complex organic molecules. | |
| cd03300 | ATP-binding cassette domain of the polyamine transporter. | PotA is an ABC-type transporter and the ATPase component of the spermidine/putrescine-preferential uptake system consisting of PotA, -B, -C, and -D. PotA has two domains with the N-terminal domain containing the ATPase activity and the residues required for homodimerization with PotA and heterdimerization with PotB. | |
| COG1127 | ABC-type transporter Mla maintaining outer membrane lipid asymmetry, ATPase component MlaF. | Cell wall/membrane/envelope biogenesis. | |
| cd03369 | ATP-binding cassette domain 2 of NFT1, subfamily C. | Domain 2 of NFT1 (new full-length MRP-type transporter 1). NFT1 belongs to the MRP (multidrug resistance-associated protein) family of ABC transporters. | |
| cd02079 | P-type heavy metal-transporting ATPase. | Heavy metal-transporting ATPases (Type IB ATPases) transport heavy metal ions (Cu(+), Cu(2+), Zn(2+), Cd(2+), Co(2+), etc.) across biological membranes. | |
| cd00994 | Glutamine binding domain of ABC-type transporter; the type 2 periplasmic binding protein fold. | This periplasmic substrate-binding component serves as an initial receptor in the ABC transport of glutamine. GlnH belongs to the type 2 periplasmic-binding fold protein (PBP2) superfamily, whose members are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. | |
| cd02076 | Plant and fungal plasma membrane H(+)-ATPases. | This subfamily includes eukaryotic plasma membrane H(+)-ATPase which transports H(+) from the cytosol to the extracellular space, thus energizing the plasma membrane for the uptake of ions and nutrients. | |
| cd01431 | ATP-dependent membrane-bound cation and aminophospholipid transporters. | The P-type ATPases, are a large family of integral membrane transporters that are of critical importance in all kingdoms of life. They generate and maintain (electro-) chemical gradients across cellular membranes, by translocating cations, heavy metals and lipids. | |
| cd03227 | ATP-binding cassette domain of non-transporter proteins. | ABC-type class 2 contains systems involved in cellular processes other than transport. These families are characterized by the fact that the ABC subunit is made up of duplicated, fused ABC modules (ABC2). | |
| cd06340 cd06349 | ABC transporter periplasmic binding domain, type 1 (ATPase Binding Cassette). | This subgroup includes the type 1 periplasmic ligand-binding domain of uncharacterized ABC (ATPase Binding Cassette)-type active transport systems that are predicted to be involved in transport of amino acids, peptides, or inorganic ions. | |
| Defense plant pathogens | cd05489 | TAXI-I inhibits degradation of xylan in the cell wall. | Xylanase inhibitor-I (TAXI-I) is a member of potent TAXI-type inhibitors of fungal and bacterial family 11 xylanases. Plants developed a diverse battery of defense mechanisms in response to continual challenges by a broad spectrum of pathogenic microorganisms. Their defense arsenal includes inhibitors of cell wall-degrading enzymes, which hinder a possible invasion and colonization by antagonists. |
| cd01960 | nsLTP1: Non-specific lipid-transfer protein type 1 (nsLTP1) subfamily. | Plant nsLTPs are small, soluble proteins that facilitate the transfer of fatty acids, phospholipids, glycolipids, and steroids between membranes. In addition to lipid transport and assembly, nsLTPs also play a key role in the defense of plants against pathogens. They may be involved in the formation of cutin layers on plant surfaces by transporting cutin monomers. | |
| cd01751 | LAT/ LH2 domain of plant lipoxygenase related proteins. | Lipoxygenases are nonheme, nonsulfur iron dioxygenases that act on lipid substrates containing one or more (Z,Z)-1,4-pentadiene moieties. In plants, the immediate products are involved in defense mechanisms against pathogens and may be precursors of metabolic regulators. The generally proposed function of PLAT/LH2 domains is to mediate interaction with lipids or membrane bound proteins. | |
| smart01037 | Pathogenesis-related protein Bet v I family. | This family is named after Bet v 1, the major birch pollen allergen. This protein belongs to family 10 of plant pathogenesis-related proteins (PR-10), cytoplasmic proteins of 15-17 kd that are wide-spread among dicotyledonous plants (PUBMED:9417891). |
This study included the search for conserved protein domain families involved in defense against plant pathogens. Within this functional group, we identified xylanase inhibitor I (TAXI-I) -a member of the potent TAXI-type inhibitors of fungal and bacterial family 11 xylanases (cd05489), plant nsLTP proteins facilitating the transfer of fatty acids, phospholipids, glycolipids, and steroids between membranes and participating in the formation of cutin layers on the surface of plants through the transport of cutin monomers, which play a key role in the defense of plants against pathogens (cd01960), and the LAT/LH2 domain of lipoxygenase related proteins -their immediate products in plants being involved in defense mechanisms against pathogens and that might be precursors of metabolic regulators (cd01751).
Enzymes involved in the secondary metabolism of species in the family Cactaceae in relation to species in other plant classes and families
The results of our analysis of the evolutionary relationships of enzymes involved in the secondary metabolism in the analyzed genomes of members of the Cactaceae family showed high conservation of the aspartic proteinase-like protein (Fig. 2), flavanone 3-hydroxylase (F3H) (Fig. 3), hydroxycinnamoyl-CoA shikimate quinate hydroxycinnamoyl transferase (HCT) (Fig. 4), serine threonine-protein phosphatase (Fig. 5) in Cereus fernambucensis (JALPLW01.1), Selenicereus undatus (JACYFF01.1), Stenocereus thurberi (NCQT01.1), Lophocereus schottii (NCQV01.1), Pachycereus pringlei (NCQS01.1), Pereskia humboldtii (NCQU01.1), and Carnegiea gigantea (NCQR01.1). In Cereus fernambucensis, we found divergence in PEN3 (Fig. 6) and the absence of sterol methyltransferase (SMT1; Fig. 7). To understand the conservation of the enzyme flavanone 3-hydroxylase (F3H) (Fig. 3), we modeled the three-dimensional structures of aa translated sequences we identified in the genomes of cacti and compared them with those of other plant accessions (Fig. 8). The three-dimensional modeling of a dimer created by means of a consensus sequence of the regions we obtained in our analysis of the genomes of species in the Cactaceae family (Fig. 8(B)), and its comparison with the reference model 1gp4.1A anthocyanidin synthase described in Arabidopsis thaliana (Fig. 8(A)) [34], indicated 88 % identity relative to the reference model (Fig. 8(A) - 8(B)). The tertiary structure modeling including 10 plant accessions -Gossypium arboreum (XP_017638014.1), Vernicia fordii (ARV78456.1), Hibiscus sabdariffa (ALB35017.1), Durio zibethinus (XP_022741107.1), Althaea officinalis (UOI87842.1), Dimocarpus longan (ABO48521.1), Arachis hypogaea (XP_025694592.1), Boehmeria nivea (QBC98316.1), Ziziphus jujuba (XP_015889639.1), and Litchi chinensis (ADO95201.1)- showed 30 % similarity relative to the reference model (Fig. 8 C-L). In none of the models generated in the plant accessions was it possible to identify the binding sites for 2-oxoglutaric acid (AKG) and 2-(N-morpholino)-ethanesulfonic acid (MES). However, where the domain corresponding to the 2OG-Fe (II) oxygenase superfamily was identified, a region of 136 aa was conserved between the analyzed plant accessions (Fig. 8(M)). The 2-oxoglutarate iron-dependent domain defines many classes of flavonoid enzymes. Enzymes with 2-oxoglutarate (2OG) and Fe (II)-dependent oxygenase domains include the C-terminal of prolyl 4-hydroxylase alpha subunit. The holoenzyme has the activity EC:1.14.11.2 oxygenase mixed function that catalyzes the hydroxylation of a prolyl-glycyl-containing peptide, usually in protocollagen, to a hydroxyprolyl-glycyl-containing peptide. The enzyme utilizes molecular oxygen with a concomitant oxidative decarboxylation of 2-oxoglutaramate to succinate. The full enzyme consists of an alpha2-beta2 complex with the alpha subunit contributing most of the parts of the active site. The family also includes lysyl hydrolases, isopenicillin synthases and AlkB.
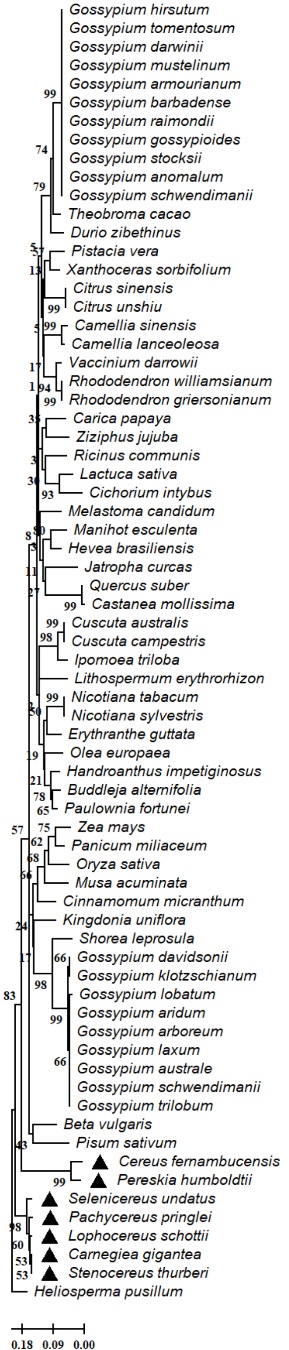
Fig. 2 Evolutionary relationships of the aspartic proteinase-like partial protein of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 139 amino acid residues from the 70 accessions Carnegiea gigantea (NCQR01056339.1), Cereus fernambucensis (JALPLW010221723.1), Lophocereus schottii (NCQV01146979.1), Pachycereus pringlei (NCQS01104914.1), Pereskia humboldtii (NCQU01061983.1), Selenicereus undatus (JACYFF010031958.1), Stenocereus thurberi (NCQT01038052.1), Heliosperma pusillum (KAH9605552.1), Camellia sinensis (XP_028103656.1), Beta vulgaris (XP_010679567.1), Shorea leprosula (GKV10306.1), Jatropha curcas (KDP32026.1), Kingdonia uniflora (KAF6142399.1), Melastoma candidum (KAI4330257.1), Gossypium raimondii (MBA0589271.1), Gossypium aridum (MBA0676517.1), Gossypium barbadense (PPD68450.1), Gossypium arboreum (XP_017620942.1), Gossypium armourianum (MBA0831389.1), Gossypium lobatum (MBA0550510.1), Gossypium mustelinum (TYI54146.1), Gossypium darwinii (TYG43918.1), Gossypium davidsonii (MBA0607055.1), Gossypium hirsutum (XP_016731018.1), Gossypium tomentosum (TYH42324.1), Gossypium laxum (MBA0705948.1), Gossypium gossypioides (MBA0741103.1), Gossypium stocksii (KAH1080006.1), Gossypium australe (KAA3466933.1), Gossypium anomalum (KAG8477069.1), Gossypium schwendimanii (MBA0849879.1), Gossypium trilobum (MBA0759655.1), Gossypium klotzschianum (MBA0642009.1), Manihot esculenta (KAG8641174.1), Heliosperma pusillum (KAH9603065.1), Carica papaya (XP_021908155.1), Pisum sativum (KAI5439377.1), Lactuca sativa (XP_023754333.1), Olea europaea (XP_022875741.1), Shorea leprosula (GKV10307.1), Ricinus communis (EEF37449.1), Hevea brasiliensis (XP_021635328.1), Quercus suber (POE98796.1), Vaccinium darrowii (KAH7850984.1), Nicotiana tabacum (XP_016495754.1), Castanea mollissima (KAF3966220.1), Shorea leprosula (GKV04501.1), Pistacia vera (XP_031277948.1), Lithospermum erythrorhizon (KAG9151776.1), Theobroma cacao (XP_007047435.2), Cuscuta australis (RAL52469.1), Cuscuta campestris (VFQ69060.1), Handroanthus impetiginosus (PIN24030.1), Buddleja alternifolia (KAG8363793.1), Oryza sativa (KAB8106186.1), Citrus sinensis (KAH9712172.1), Citrus unshiu (GAY50237.1), Cinnamomum micranthum (RWR78019.1), Camellia lanceoleosa (KAI8018936.1), Musa acuminata (XP_009395627.1), Rhododendron williamsianum (KAE9466797.1), Paulownia fortunei (KAI3448654.1), Rhododendron griersonianum (KAG5535551.1), Ziziphus jujuba (KAH7520274.1), Ziziphus jujuba (XP_015901624.1), Ipomoea triloba (XP_031102984.1), Jatropha curcas (XP_012079339.1), Durio zibethinus (XP_022741477.1), Xanthoceras sorbifolium (KAH7566657.1), Cichorium intybus (KAI3722646.1), Zea mays (ONM59313.1), Erythranthe guttata (EYU43423.1), Nicotiana sylvestris (XP_009780676.1) and Panicum miliaceum (RLM85991.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
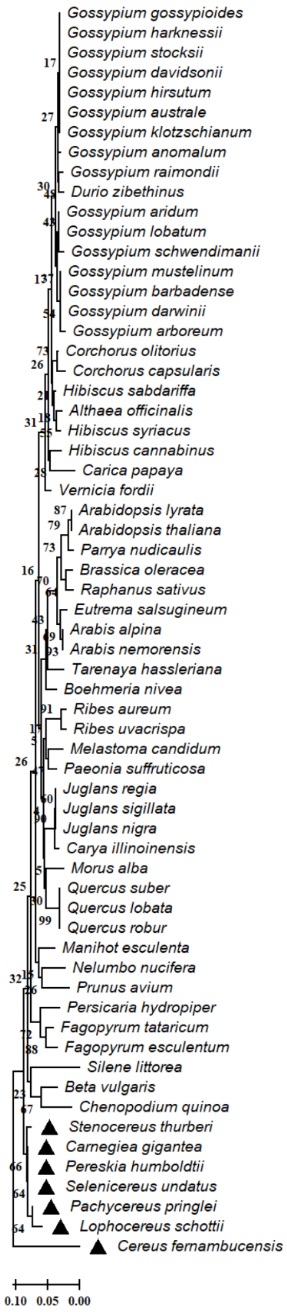
Fig. 3 Evolutionary relationships of the flavanone 3-hydroxylase (F3H) partial enzyme of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 62 amino acid residues from the 71 accessions Carnegiea gigantea (NCQR01052635.1), Cereus fernambucensis (JALPLW010088659.1), Stenocereus thurberi (NCQT01019950.1), Pachycereus pringlei (NCQS01148615.1), Lophocereus schottii (NCQV01146979.1), Pereskia humboldtii (NCQU01107586.1), Selenicereus undatus (JACYFF010031956.1), Beta vulgaris (KMT09612.1), Ribes aureum (CEL26663.1), Vernicia fordii (ARV78456.1), Hibiscus sabdariffa (ALB35017.1), Tarenaya hassleriana (XP 010550464.1), Eutrema salsugineum (XP 006403939.1), Corchorus olitorius (OMO84632.1), Melastoma candidum (KAI4325433.1), Althaea officinalis (UOI87842.1), Boehmeria nivea (QBC98316.1), Fagopyrum tataricum (AEF30420.1), Hibiscus syriacus (XP 039020374.1), Gossypium klotzschianum (MBA0655530.1), Persicaria hydropiper (BAB85681.1), Paeonia suffruticosa (AEN71544.1), Hibiscus cannabinus (AHY24651.1), Gossypium aridum (MBA0688295.1), Arabidopsis lyrata (XP 020879934.1), Juglans nigra (CAB97360.1), Gossypium australe (KAA3461107.1), Gossypium hirsutum (NP 001314423.1), Gossypium davidsonii (MBA0620121.1), Gossypium raimondii (KJB48219.1), Juglans regia (XP 018829624.1), Juglans sigillata (AYK27186.1), Gossypium stocksii (KAH1072761.1), Arabidopsis thaliana (NP 001190050.1), Gossypium gossypioides (MBA0753123.1), Arabis alpina (KFK34366.1), Ribes uvacrispa (CEL26665.1), Morus alba (AOV62761.1), Carya illinoinensis (XP 042988041.1), Fagopyrum esculentum (ADT63064.1), Nelumbo nucifera (XP 010268217.1), Silene littorea (AMQ23621.1), Gossypium anomalum (KAG8474570.1), Gossypium harknessii (MBA0804691.1), Gossypium lobatum (MBA0562421.1), Carica papaya (XP 021908198.1), Manihot esculenta (XP 021605137.1), Arabis nemorensis (VVB05321.1), Durio zibethinus (XP 022741107.1), Gossypium mustelinum (TYJ03983.1), Gossypium barbadense (KAB2051536.1), Gossypium darwinii (TYG88995.1), Chenopodium quinoa (XP 021727556.1), Brassica oleracea (XP 013599994.1), Quercus suber (XP 023924008.1), Quercus lobata (XP 030975239.1), Quercus robur (XP 050292388.1), Corchorus capsularis (OMO57049.1), Gossypium schwendimanii (MBA0862352.1), Raphanus sativus (BAC58033.1), Parrya nudicaulis (ADY02696.1), Gossypium arboreum (XP 017638014.1) and Prunus avium (XP 021815794.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
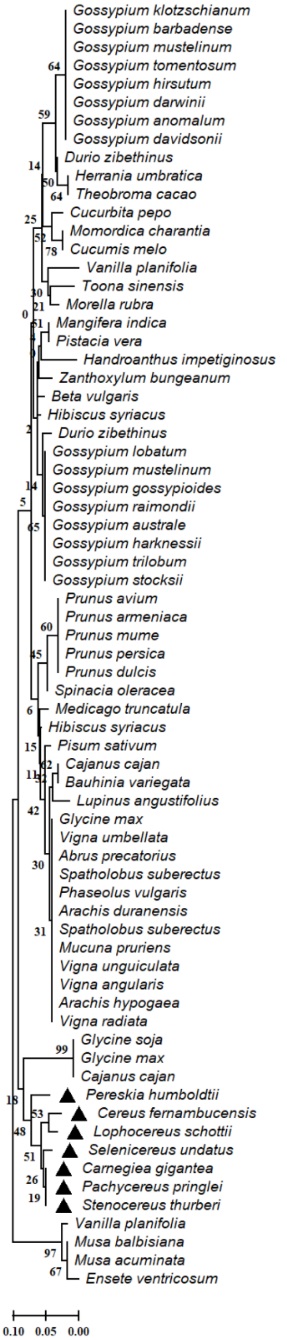
Fig. 4 Evolutionary relationships of the hydroxycinnamoyl-CoA shikimate quinate hydroxycinnamoyl transferase (HCT) partial enzyme of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 62 amino acid residues from the 70 accessions Carnegiea gigantea (NCQR01003266.1), Cereus fernambucensis (JALPLW010256838.1), Lophocereus schottii (NCQV01118460.1), Pachycereus pringlei (NCQS01005339.1), Pereskia humboldtii (NCQU01069994.1), Selenicereus undatus (JACYFF010002017.1), Stenocereus thurberi (NCQT01149583.1), Prunus dulcis (XP 034215172.1), Prunus persica (XP 020419855.1), Durio zibethinus (XP 022733842.1), Beta vulgaris (XP 010695048.2), Gossypium davidsonii (MBA0610884.1), Spinacia oleracea (XP 021846761.1), Toona sinensis (QWM97876.1), Vanilla planifolia (KAG0473240.1), Lupinus angustifolius (XP 019459362.1), Cajanus cajan (XP 020227010.1), Hibiscus syriacus (XP 039062574.1), Glycine max (BBL33356.1), Herrania umbratica (XP 021276713.1), Vigna umbellata (XP 047179389.1), Durio zibethinus (XP 022773545.1), Medicago truncatula (XP 013444592.1), Musa balbisiana (THU53406.1), Cajanus cajan (XP 020216908.1), Momordica charantia (XP 022158366.1), Abrus precatorius (XP 027334676.1), Spatholobus suberectus (TKY64105.1), Pisum sativum (KAI5423757.1), Cucurbita pepo (XP 023520534.1), Prunus mume (XP 008239196.1), Theobroma cacao (EOY23952.1), Phaseolus vulgaris (XP 007135274.1), Mangifera indica (XP 044493298.1), Arachis duranensis (XP 015968240.1), Pistacia vera (XP 031253990.1), Spatholobus suberectus (TKY61240.1), Hibiscus syriacus (KAE8700978.1), Gossypium lobatum (MBA0562932.1), Mucuna pruriens (RDX66809.1), Morella rubra (UVJ68634.1), Gossypium mustelinum (TYI50838.1), Gossypium gossypioides (MBA0743674.1), Gossypium anomalum (KAG8486051.1), Vanilla planifolia (KAG0499750.1), Gossypium darwinii (TYH06026.1), Glycine soja (XP 028215100.1), Gossypium hirsutum (XP 016738141.1), Prunus avium (XP 021816604.1), Bauhinia variegata (KAI4328937.1), Zanthoxylum bungeanum (URY18891.1), Gossypium raimondii (XP 012437779.1), Gossypium australe (KAA3462300.1), Vigna unguiculata (XP 027932607.1), Gossypium tomentosum (TYI14493.1), Vigna angularis (XP 017435245.1), Gossypium harknessii (MBA0805237.1), Musa acuminata (XP 009379853.1), Prunus armeniaca (CAB4280244.1), Arachis hypogaea (XP 025657499.1), Gossypium mustelinum (TYI69172.1), Cucumis melo (KAA0042662.1), Glycine max (XP 003516658.1), Gossypium trilobum (MBA0772507.1), Gossypium stocksii (KAH1073765.1), Gossypium klotzschianum (MBA0645960.1), Handroanthus impetiginosus (PIN00955.1), Gossypium barbadense (KAB2017041.1), Vigna radiata (XP 014491519.1) and Ensete ventricosum (RWW34505.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
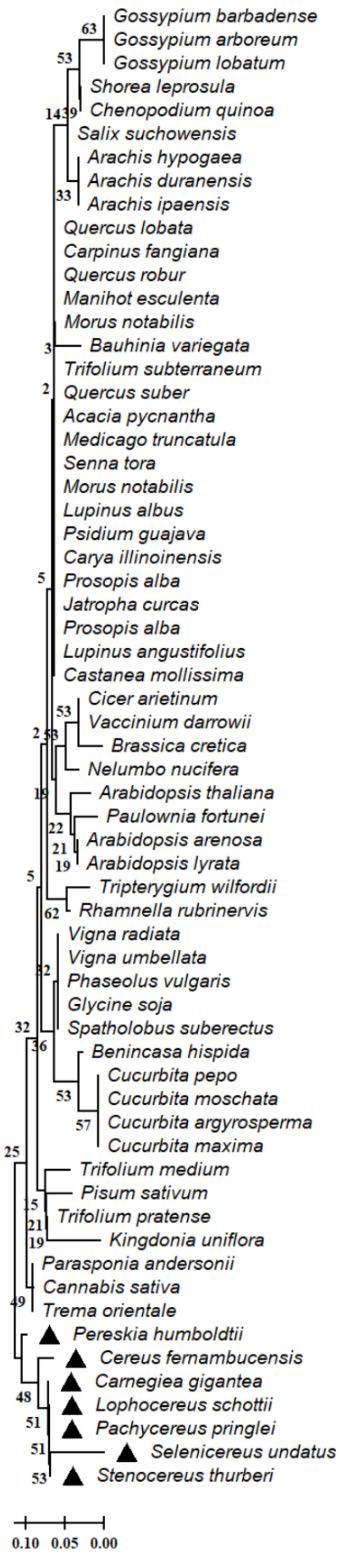
Fig. 5 Dendrogram of the serine threonine-protein phosphatase 7 partial of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 32 amino acid residues from the 63 accessions Cereus fernambucensis (JALPLW010228804.1), Carnegiea gigantea (NCQR01000535.1), Lophocereus schottii (NCQV01152660.1), Pachycereus pringlei (NCQS01079590.1), Pereskia humboldtii (NCQU01013109.1), Selenicereus undatus (JACYFF010031957.1), Stenocereus thurberi (NCQT01097179.1), Trifolium medium (MCI27547.1), Trifolium pratense (XP 045808829.1), Carya illinoinensis (KAG6647577.1), Cannabis sativa (KAF4370178.1), Morus notabilis (EXB64215.1), Psidium guajava (KAI3423248.1), Prosopis alba (XP 028762166.1), Medicago truncatula (XP 003590590.1), Senna tora (KAF7818353.1), Jatropha curcas (KDP21412.1), Lupinus albus (KAE9618420.1), Acacia pycnantha (KAI9119432.1), Prosopis alba (XP 028762168.1), Quercus suber (POF06493.1), Trifolium subterraneum (GAU35616.1), Lupinus angustifolius (XP 019426578.1), Carpinus fangiana (KAE8009768.1), Quercus lobata (XP 030946147.1), Castanea mollissima (KAF3956392.1), Quercus robur (XP 050264930.1), Manihot esculenta (XP 021610441.1), Morus notabilis (XP 024021424.1), Spatholobus suberectus (TKY52655.1), Glycine soja (XP 028206358.1), Phaseolus vulgaris (XP 007144658.1), Cucurbita pepo (XP 023513670.1), Cucurbita moschata (XP 022959987.1), Cucurbita argyrosperma (KAG6593098.1), Cicer arietinum (XP 004495174.1), Arachis hypogaea (XP 025642825.1), Arachis duranensis (XP 015948558.1), Pisum sativum (KAI5395123.1), Bauhinia variegata (KAI4349474.1), Arachis ipaensis (XP 016182894.1), Cucurbita maxima (XP 023004800.1), Vigna radiata (XP 014512427.1), Brassica cretica (KAF2605352.1), Vaccinium darrowii (KAH7864243.1), Vigna umbellata (XP 047175158.1), Arabidopsis arenosa (CAE6081884.1), Shorea leprosula (GKU86520.1), Arabidopsis thaliana (NP 191888.2), Parasponia andersonii (PON54453.1), Gossypium lobatum (MBA0561982.1), Tripterygium wilfordii (KAF5727048.1), Salix suchowensis (KAG5250564.1), Rhamnella rubrinervis (KAF3436257.1), Trema orientale (PON94958.1), Benincasa hispida (XP 038897661.1), Paulownia fortunei (KAI3449677.1), Chenopodium quinoa (XP 021770304.1), Gossypium barbadense (KAB1997119.1), Arabidopsis lyrata (XP 020882119.1), Nelumbo nucifera (XP 010259132.1), Gossypium arboreum (XP 017612360.1) and Kingdonia uniflora (KAF6177019.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
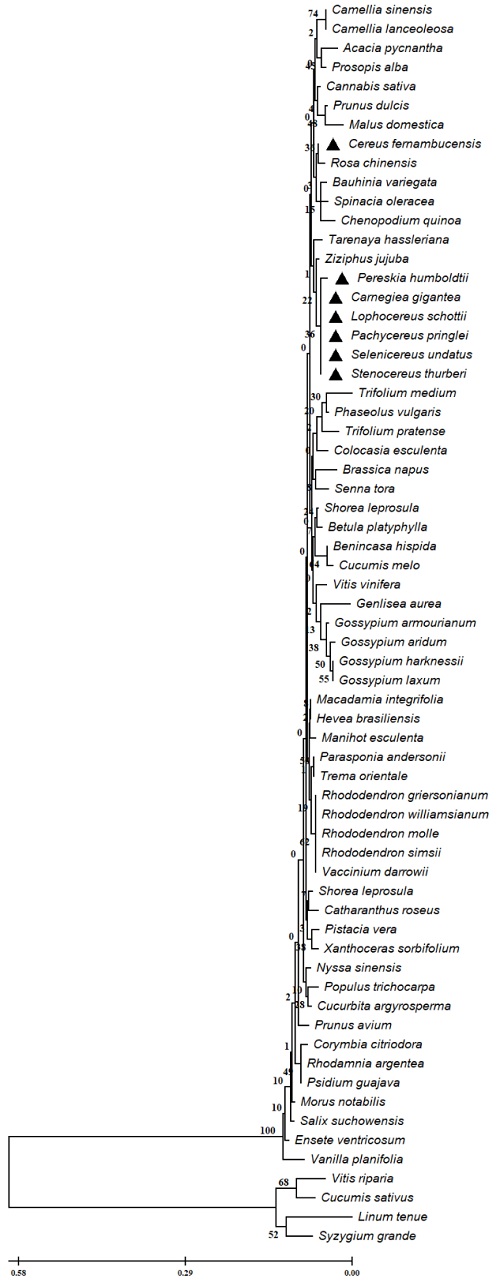
Fig. 6 Dendrogram of the PDR ABC-type transporter family protein (PEN3) partial of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 88 amino acid residues from the 65 accessions Carnegiea gigantea (NCQR01026447.1), Cereus fernambucensis (JALPLW010367148.1), Lophocereus schottii (NCQV01109799.1), Pachycereus pringlei (NCQS01144854.1), Pereskia humboldtii (NCQU01045749.1), Selenicereus undatus (JACYFF010031955.1), Stenocereus thurberi (NCQT01156589.1), Camellia sinensis (XP 028063537.1), Vitis vinifera (CAN76184.1), Acacia pycnantha (KAI9109544.1), Trifolium medium (MCH87329.1), Trifolium pratense (PNX77073.1), Prunus dulcis (VVA18642.1), Vitis riparia (XP 034695469.1), Ensete ventricosum (RWW84675.1), Macadamia integrifolia (XP 042517769.1), Prunus avium (XP 021818782.1), Bauhinia variegata (KAI4355588.1), Camellia lanceoleosa (KAI7986309.1), Ziziphus jujuba (XP 048323260.1), Rhodamnia argentea (XP 030525586.2), Parasponia andersonii (PON54345.1), Trema orientale (PON88559.1), Spinacia oleracea (XP 021853414.1), Cannabis sativa (KAF4362150.1), Shorea leprosula (GKV36634.1), Chenopodium quinoa (XP 021765340.1), Rhododendron griersonianum (KAG5537773.1), Hevea brasiliensis (XP 021659471.1), Linum tenue (CAI0423367.1), Populus trichocarpa (KAI5602673.1), Shorea leprosula (GKV27175.1), Rhododendron williamsianum (KAE9467878.1), Nyssa sinensis (KAA8540358.1), Brassica napus (CDY69529.1), Rhododendron molle (KAI8544504.1), Psidium guajava (KAI3416940.1), Gossypium armourianum (MBA0840459.1), Tarenaya hassleriana (XP 010528954.1), Senna tora (KAF7803604.1), Syzygium grande (KAI6690085.1), Gossypium harknessii (MBA0812280.1), Gossypium laxum (MBA0724446.1), Cucurbita argyrosperma (KAG6598625.1), Benincasa hispida (XP 038896688.1), Catharanthus roseus (KAI5651241.1), Colocasia esculenta (MQL78829.1), Corymbia citriodora (KAF8025632.1), Malus domestica (RXH88665.1), Rhododendron simsii (KAF7135728.1), Manihot esculenta (XP 021610472.1), Pistacia vera (XP 031255754.1), Morus notabilis (XP 010098138.1), Betula platyphylla (QMS43712.1), Vaccinium darrowii (KAH7865069.1), Cucumis sativus (NP 001292689.1), Cucumis melo (XP 008447550.2), Genlisea aurea (EPS70689.1), Phaseolus vulgaris (XP 007150610.1), Xanthoceras sorbifolium (KAH7565305.1), Vanilla planifolia (KAG0499329.1), Rosa chinensis (XP 024184027.1), Gossypium aridum (MBA0695701.1), Prosopis alba (XP 028783605.1) and Salix suchowensis (KAG5229136.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
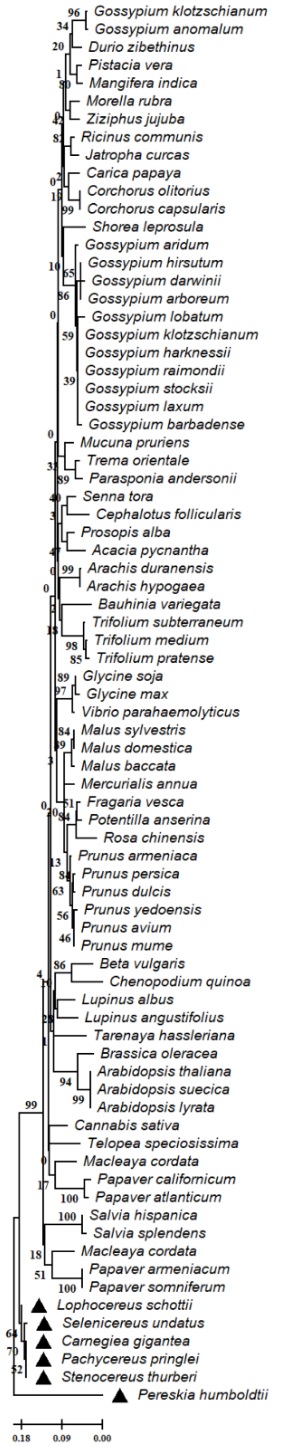
Fig. 7 Dendrogram of the sterol methytransferase (SMT1) protein partial of Cactaceae and plant species. The evolutionary relationships were performed using the MEGA 11 program with the minimal evolution method and with a bootstrap of 1000 repetitions. Partial sequences of 131 amino acid residues from the 78 accessions Carnegiea gigantea (NCQR01000865.1), Lophocereus schottii (NCQV01032762.1), Pachycereus pringlei (NCQS01134798.1), Pereskia humboldtii (CQU01116023.1), Selenicereus undatus (JACYFF010019584.1), Stenocereus thurberi (NCQT01031414.1), Fragaria vesca (XP 004291325.1), Vibrio parahaemolyticus (WP 169679238.1), Potentilla anserina (XP 050363276.1), Macleaya cordata (OVA15971.1), Trema orientale (PON93969.1), Corchorus olitorius (OMO75464.1), Malus baccata (TQE07532.1), Gossypium aridum (MBA0691512.1), Parasponia andersonii (PON34897.1), Malus sylvestris (XP 050158836.1), Malus domestica (XP 008377126.2), Macleaya cordata (OVA08873.1), Beta vulgaris (XP 010667472.1), Corchorus capsularis (OMP12330.1), Trifolium medium (MCH98114.1), Pistacia vera (XP 031265443.1), Glycine soja (KAG5018186.1), Lupinus albus (KAF1893465.1), Prunus armeniaca (KAH0990658.1), Tarenaya hassleriana (XP 010534003.1), Senna tora (KAF7829524.1), Cannabis sativa (XP 030507667.1), Cephalotus follicularis (GAV75510.1), Mangifera indica (XP 044474559.1), Prosopis alba (XP 028765915.1), Trifolium pratense (XP 045806382.1), Salvia hispanica (XP 047941495.1), Acacia pycnantha (KAI9113296.1), Gossypium arboreum (XP 017621279.1), Gossypium klotzschianum (MBA0659018.1), Gossypium hirsutum (XP 016725967.1), Arachis duranensis (XP 015933966.1), Gossypium harknessii (MBA0818334.1), Gossypium raimondii (XP 012445831.1), Gossypium stocksii (KAH1065840.1), Gossypium laxum (MBA0720516.1), Arachis hypogaea (RYR41286.1), Ricinus communis (XP 002510600.1), Prunus avium (XP 021827540.1), Mucuna pruriens (RDX70222.1), Gossypium darwinii (TYH17479.1), Morella rubra (KAB1203606.1), Prunus mume (XP 008221201.1), Carica papaya (XP 021902736.1), Salvia splendens (XP 042004281.1), Jatropha curcas (XP 012073720.1), Brassica oleracea (XP 013615337.1), Ziziphus jujuba (XP 015887788.1), Prunus persica (XP 020423207.1), Prunus dulcis (KAI5353641.1), Papaver armeniacum (KAI3925018.1), Trifolium subterraneum (GAU24946.1), Mercurialis annua (XP 050212443.1) , Shorea leprosula (GKV17786.1), Papaver somniferum (XP 026396855.1), Gossypium klotzschianum (MBA0659640.1), Glycine max (ACS93764.1), Lupinus angustifolius (XP 019416658.1), Chenopodium quinoa (XP 021756554.1), Prunus yedoensis (PQQ17669.1), Gossypium barbadense (KAB2029880.1), Durio zibethinus (XP 022772608.1), Gossypium lobatum (MBA0565757.1), Gossypium anomalum (KAG8493703.1), Arabidopsis thaliana (NP 173458.1), Rosa chinensis (XP 024192147.1), Arabidopsis suecica (KAG7654950.1), Bauhinia variegata (KAI4336732.1), Papaver californicum (KAI3980587.1), Papaver atlanticum (KAI3916092.1), Arabidopsis lyrata (XP 020868655.1) and Telopea speciosissima (XP 043708782.1). The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
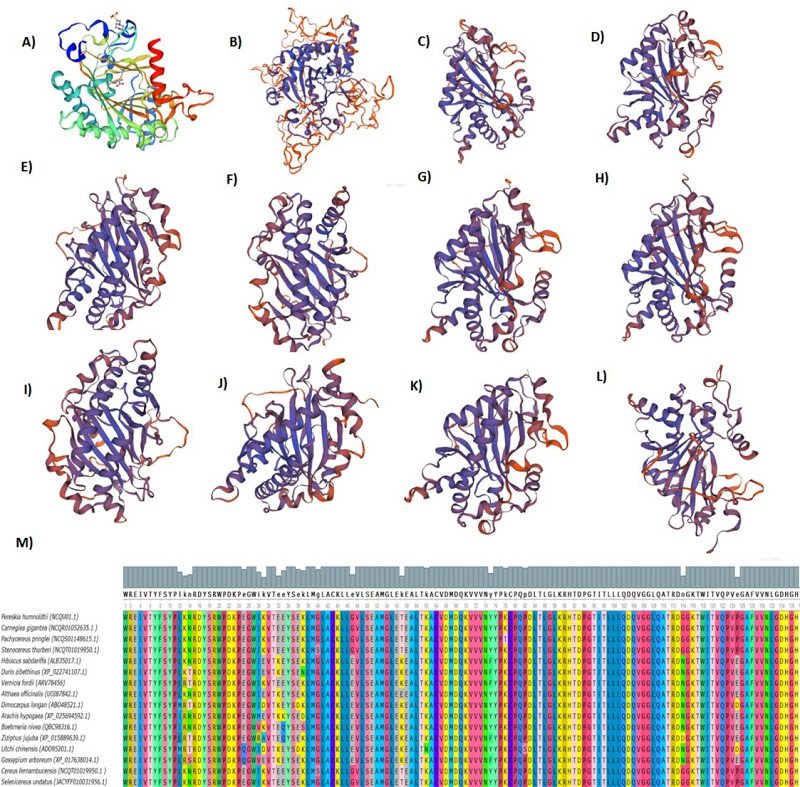
Fig. 8 In silico tertiary structures of flavanone 3-hydroxylase (F3H)) in Cactus spp. The prediction of tertiary conformation was performed using the SWISSMODEL program and Arabidopsis thaliana was used as a reference. Predicted three-dimensional structures of (A) Arabidopsis thaliana reference (1gp4.1A), (B) Cactus consensus sequence-dimer, (C) Gossypium arboreum (XP_017638014.1), (D) Vernicia fordii (ARV78456.1), (E) Hibiscus sabdariffa (ALB35017.1), (F) Durio zibethinus (XP_022741107.1) (G) Althaea officinalis (UOI87842.1), (H) Dimocarpus longan (ABO48521.1), (I) Arachis hypogaea (XP_025694592.1), (J) Boehmeria nivea (QBC98316.1), (K) Ziziphus jujuba (XP_015889639.1) and (L) Litchi chinensis (ADO95201.1). (M) Alignment of a large region of the α2-oxoglutarate iron-dependent domain in 16 plant accessions. The analysis of similarity of amino acid residues was carried out with 10 classes of plants and 6 species of cataceae: Pereskia humboldtii (NCQU01.1), Carnegiea gigantea (NCQR01052635.1), Pachycereus pringlei (NCQS01148615.1), Stenocereus thurberi (NCQT019950. 1), Cereus fenambucensis (NCQT01019950.1) and Selenecereus undatus (JACYFF0100319561.1) with which the cactus dimer was constructed. The line indicates the scale of the genetic distance of the sequences and the numbers the bootstrap support and confidence of resolution of each separation.
As we demonstrated in this study, even though a large region of the 2-oxoglutarate iron-dependent domain proved to be conserved among the analyzed plant accessions, modeling of their three-dimensional structures indicated changes in the aa sequences of the a-binding 2-oxoglutaric acid (AKG) and 2-(N-morpholino)-ethanesulfonic acid (MES), possibly indicating point mutations affecting these aa residues. These enzymes are in constant evolution and perhaps depending on the secondary metabolism capabilities of each plant species, it leads to setting mutations that favor enzyme specialization [43-45]. Studies directed at identifying specific changes in these secondary metabolism enzymes might be a near-term project that would allow us to use these enzymes in genetic engineering for improving flavonoid production in plants.
Conclusion
Obtaining bioactive compounds from plants and their application in the food industry and medicine is a reality today. Species belonging to the family Cactaceae could be used to extract different bioactive compounds and the results of bioinformatic analyses would allow us to advance in the search for enzymes related to secondary metabolism. However, little progress exists in the sequencing of genomes of members belonging to the family Cactaceae. In this study we were able to demonstrate that using bioinformatics tools it is possible to select candidate genes that participate in secondary metabolism and, in the future, use them to carry out genetic improvement in plants aimed at enhancing the yields of active compounds of interest.











 nueva página del texto (beta)
nueva página del texto (beta)


