Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias pecuarias
versión On-line ISSN 2448-6698versión impresa ISSN 2007-1124
Rev. mex. de cienc. pecuarias vol.7 no.1 Mérida ene./mar. 2016
Technical notes
Adaptation of Mycobacterium smegmatis to nutrient depletion and its effect on esat-6 expression
a Facultad de Medicina Veterinaria de la Universidad Autónoma de Sinaloa, Boulevard San Ángel S/N, Fraccionamiento San Benito, Predio las Coloradas, Culiacán, Sinaloa, México.
b Instituto Nacional de Investigaciones Forestales Agrícolas y Pecuarias. México.
c Facultad de Medicina Veterinaria y Zootecnia de la Benemérita Universidad Autónoma de Puebla. México.
d Centro de Innovación y Desarrollo Educativo. México.
When resources are scarce, mycobacteria stop growing to make way for genes adaptation allow. Conversely, when growth continues under stress conditions, specific genes metabolic networks for protection are activating. In this sense, the protein encoded by esat-6 (early secretory antigenic target, 6 kDa) gene in Mycobacterium tuberculosis, acting in the lysis of alveolar epithelial and macrophage membranes to escape and invade other cells. But it can have other functions, such as interfering with cell-cell contact and transfer their DNA. In M. smegmatis, the ESX-1 (Secretion Ejectosoma BOX) system that facilitates secretion of ESAT-6 protein, probably it is sensitive to one or more nutrients of the culture medium. So it in the present study culture conditions limiting nutrient for growth of M. smegmatis are evaluated and relate the expression of esat-6. Culture media tested were minimal medium Hartmans Bond (HdB), carbon-limited (HdB<C), nitrogen (HdB<N) and inorganic phosphate (HdB<Pi). M. smegmatis HdB adapted to minimal medium, HdB<C and HdB<N and resume its metabolic activity in fresh medium, but not expressed esat-6. In HdB <Pi M. smegmatis loses its metabolic capacity for resistance acid-alcohol and expressed esat-6. Therefore, the culture media tested as a model for gene expression under nutrient limitation is proposed.
Keywords: Metabolism; Dormancy; Adaptation; Stress; Nutrient depletion; Mycobacterium smegmatis; esat-6
Cuando los recursos son escasos, las micobacterias detienen su crecimiento para dar paso a los genes de la adaptación. Contrariamente, cuando el crecimiento continúa bajo condiciones de estrés, se activan genes específicos de redes metabólicas para su protección. En este sentido, la proteína codificada por esat-6 (por sus siglas en inglés: early secretory antigenic target, 6 kDa) en Mycobacterium tuberculosis, actúa en la lisis del epitelio alveolar y membranas de los macrófagos para escapar e invadir otras células. Pero puede tener otras funciones, tales como interferir en el contacto célula-célula y transferir su ADN. En M. smegmatis, el sistema ESX-1 (por sus siglas en inglés: Secretion Ejectosoma BOX) facilita la secreción de la proteína ESAT-6, probablemente es sensible a uno o más nutrientes del medio de cultivo. Por lo que en el presente estudio se evalúan las condiciones de cultivo limitantes en nutrientes para el crecimiento de M. smegmatis y su relación con la expresión del gen esat-6. Los medios de cultivos probados fueron Hartmans de Bond medio mínimo (HdB), limitado en carbono (HdB<C), nitrógeno (HdB<N) y fosfato inorgánico (HdB<Pi). M. smegmatis se adapta a HdB medio mínimo, HdB<C y HdB<N y reanuda su actividad metabólica en medio fresco, pero no se expresa esat-6. En HdB<Pi M. smegmatis pierde su capacidad metabólica respecto a la resistencia alcohol-ácido y expresa esat-6. Por lo tanto, se proponen los medios de cultivo probados como modelo para la expresión génica bajo limitación por nutrientes.
Palabras clave: Metabolismo; Aletargamiento; Adaptación; Estrés; Agotamiento de nutrientes; Mycobacterium smegmatis; esat-6
Mycobacterium tuberculosis complex is a genetically related group of Mycobacterium species that can cause tuberculosis. It characterizes for its ability to survive and grow within macrophages, in a stress environment provided with nutrient limitation and acidification1-3. Its adaptation takes place through mechanisms that allow to detect its environment, regulating gene expression or repression during growth and reproduction4, favoring its persistence in the host5.
The cells can respond to external stimuli by initiating a process derived from the binding of an effector to a receptor joined with the membrane. This is a transduction mechanism in which stress factors can trigger a response through gene expression, preparing the bacteria during nutrient depletion6,7. A fact that can be observed in vitro during the exponential phase and is decisive for adaptation to conditions present in the stationary phase and survival8. This involved that the cell accumulate protons from the environment to maintain pH homeostasis in the cytosol9 and polyphosphate to sustain energy10. In addition, glutamate also accumulates in M. tuberculosis and M. smegmatis11 and asparagine12, which is associated with induction dormancy of M. tuberculosis13.
The bacilli respond to stress conditions due to nutrient depletion, acid pH, and reactive oxygen and nitrogen species, caused by their growth in macrophages7,8,14. In order to respond to these stimuli, mycobacteria secrete enzymes, such as ESAT-6 and CFP-102,15, which in the cells of the host, act in necrosis processes, such as lysis of alveolar epithelium and membranes of macrophages, induction of DNA fragmentation and permeability of mitochondrial membranes15. Among the metabolic functions of mycobacteria, they regulate the transfer of DNA16.
M. smegmatis mc2155, mutant of M. smegmatis wild-type strain, is 10 to 100 times more efficiently transformed with plasmid vectors, for use in the analysis of mycobacteria function17. Because of its fast growing, it has been proposed as model for understanding the changes that promote adaptation18 and persistence of pathogenic mycobacteria19. It shares 12 out of 19 virulence genes with M. tuberculosis20, among them are esat-6 and cfp-10 genes, which encode corresponding proteins secreted by the Secretion Ejectosoma BOX21. Converse and Cox22 observed the effect of culture media in the ESX-1 secretion system, for which they suggest that this system can be sensitive to one or more culture media nutrients.
During the adaptation process, genes related to specific metabolic functions are expressed as a response to stress caused by nutrient depletion23. The adaptation of cultured mycobacteria can be observed after the stationary phase, changing to fresh culture medium where they resume their activity; therefore, in the present study, the limiting nutrient conditions for the growth of M. smegmatis and its relationship with esat-6 gene expression was evaluated.
M. smegmatis mc2155, was grown at 37 °C in Middlebrook 7H11 agar. The initial aliquots of test culture and untreated control cultures were obtained by inoculum of a M. smegmatis colony in 10 ml of liquid Middlebrook 7H9 medium and was grown for 24 h until mid-log phase (0.8-1.0 DO at 600 nm).
The culture media used as control for comparing the metabolic behavior of bacilli and expression of esat-6 gene and Sauton’s medium without zinc, both supplemented with 0.5 and 6% of glycerol, respectively, and 0.05 % of Tween 80, for being media in which esat-6 gene expresses22. The initial inoculation was carried out using 100 μl of the initial culture in 225 ml flasks with 150 ml of liquid medium and were incubated at 37 °C and agitated at 250 rpm.
All assay culture media for nutrient limitation derived from HdB minimal medium; pH 7 was adjusted and 0.2 % of glycerol and 0.05 % of Tween 80 were added, except for depletion of C (HdB<C) to which glycerol was limited to 0.08 % (v/v). For the experiments in which nitrogen depletion was required (HdB<N), (NH4)2SO4 was used at a concentration 100 times lower (0.15 mM) than the utilized for HdB minimal medium. For the experiments limited 100 times lower in Pi (HdB<Pi), K2HPO4 was added at a final concentration of 0.089 mM (0.0155 g L-1) and NaH2PO4, at a concentration of 0.0708 mM (0.085 g L-1), for replacing the loss of buffer capacity, 3-(Nmorpholino) propanesulfonic acid (MOPS) 50 mM was added24. The growth and stationary phases of all cultures were monitored at 600 nm (OD600), until 144 h and final pH was recorded.
The predicted fragment incorporating esat-6 gene was obtained by amplification of M. smegmatis genomic DNA, using primers: F-5’ACAGGTATGGAATTTCGCCG-3’, R-5’-CAGGCAAACATTCCCGTGA-3’, which were designed using the Oligo TM Software DNA Star Program. For conducting the identification by enzymatic sequencing, the amplified product was purified by a protocol based on silica columns (QIAquick Gel Extraction Kit, QIAGEN®); the amplified fragment was observed in agarose gel at 1.2 % through a UV- light transilluminator and it was cut with a sterile scalpel, carrying out the aforementioned protocol. The quality and quantity of purified DNA was observed by agarose gel and this amplified sample was sent for sequencing. This process was carried out at the Laboratorio Nacional de Genómica para la Biodiversidad (LANGEBIO) of the Centro de Investigación y Estudios Avanzados (CINVESTAV), Unidad Irapuato, Guanajuato, México, of the Instituto Politécnico Nacional (IPN), based on the ddNTPs method25, using the 3730 XL DNA sequencer (Applied Biosystems, Foster City, CA) and the kit Big Dye Terminator 3.1 (Applied Biosystems, Foster City, CA).
For RNA extraction, M. smegmatis culture was adapted to the corresponding culture media until the beginning of the stationary phase (96 h). In this phase, 30 % of cells were cultured, washed with PBS and placed in 150 mL of fresh culture medium under the same nutritional conditions and they were incubated for 14 h. RNA was extracted using TRlzol (Invitrogen®); it was observed by electrophoresis in agarose gels (1.2 %) using MOPS X (200 mM of MOPS, 50 mM of sodium acetate and 10 mM of EDTA, pH 7), and 3.15 % of formaldehyde. Electrophoresis was ran in TAE buffer solution (40/1 mM pH 8.0).
Northern blot hybridization was performed starting from 8 to 12 ng of extracted RNA, capillary transferred onto nylon membranes, sodium citrate and sodium chloride 20X. DNA was fixed using ultraviolet light for 5 min. The probe was derived from fragments of esat-6 gene, amplified by PCR and labelled as biotinylated alkaline phosphatase (Gene Images Alkphos Direct Labelling and Detection System-GE Health Care®). Hybridization was performed all night and was revealed by autoradiography (Hyperfilm-MP autoradiography, Amersham Pharmacia Bioscience®)26.
The growth and pH data were obtained from six repetitions and Student’s t- test was used to express the results of the mean, standard error of the mean and corresponding comparisons between all M. smegmatis cultures27.
In the testing culture media, M. smegmatis was observed with lower growth in HdB<N, HdB<C (0.659 and 1.697) media, in comparison to HdB minimal culture and HdB<Pi (2.170 and 1.982) (P<0.05). In addition, M. smegmatis in HdB<Pi has similar growth in HdB minimal culture (P>0.05) (Figure 1). This is indicative of a continuous growth in spite of stress conditions due to a decrease 100 times Pi. With the exception of HdB<Pi culture, in the rest of the testing and control media used, M. smegmatis resume its growth while changing it to a fresh culture medium, after 144 h of culture.
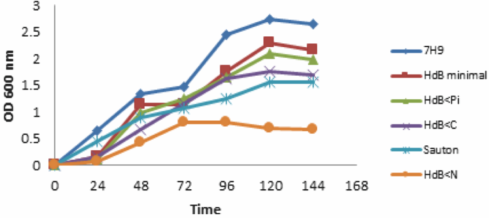
Figure 1 M. smegmatis grown until 144 h in culture media used for experiments: HdB<N (0.659±0.12), Sauton (1.559±0.03), HdB<C (1.697±0.07), HdB<Pi (1.982±0.11) HdB minimal (2.170±0.12) and 7H9 (2.638±0.12).
In HdB<Pi culture medium, a relationship between growth, loss of acid-alcohol resistance of M. smegmatis, and pH was observed. That is, by higher number of culture hours (144), pH was decreased (5.4 ± 0.16), and the mycobacteria showed incapacity to recuperate its metabolic activity when the culture was changed to a fresh medium. This was observed in cumulus cells stained with the characteristic color of Ziehl-Neelsen technique, which disappeared from 100 h, until all cells were stained blue-colored (Figures 2a and 2b).

Figure 2 M. smegmatis a) Loss of acid-alcohol resistance in HdB<Pi culture medium through 96 h. b) Same culture changed to fresh medium 1 h after, where recovery of capacity of response to acid-alcohol is observed.
With the exception of HdB<Pi culture medium, mycobacteria in limited nutrient culture media have a slow growth until they reach the stationary phase. Additionally, bacteria grown in HdB<P gradually lose their resistance to acidalcohol from 96 h, not recovering it until 144 h of culture, when bacteria were changed to fresh medium. For which the 96 h were taken into consideration for changing M. smegmatis to fresh culture media, extract total RNA after 12 ± 1 h (Figure 3A) and perform Northern hybridization. This assay reveals that M. smegmatis cultured in HdB<C, HdB<N and HdB minimal medium, does not express esat-6 gene. However, the culture limited 100 times in Pi (Figure 3B, lane 7) did showed the expression, in a similar way as the culture media used as control (7H9 and Sauton, lanes 1 and 3, Figure 3B).
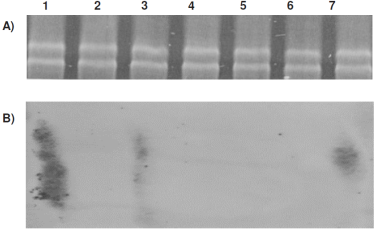
Figure 3 M. smegmatis A) Total RNA of the bacteria in culture media: 1) 7H9, 2) HdB minimal culture, 3) Sauton without zinc, 4) not controlled variable 5) HdB<N, 6) HdB<C, 7) HdB<Pi. B) Same lanes with the results of Northern hybridization for esat-6 gene.
The probe elaborated from esat-6 fragment was sequenced and was referred to GeneBank number SeqID KR363260. The homology comparison consulted on the database corresponds to esat-6 gene and is located in site 87 387 to 87 604 pb of the complete genome sequencing of M. smegmatis strain MC2155 (ID: gb|CP000480.1).
The mycobacterial capacity to accumulate energy sources, allows them to in vivo persist the infection phase28. In this sense, in vitro mycobacteria achieve to adapt to nutrient depletion and survive for more than 650 d24, due to energy accumulation of the stored compounds and degradation of unnecessary proteins and bacterial RNA6. This can cause an emergency situation, where mycobacteria respond to the environment through genes which regulate adequate changes to the conditions imposed. Thus, the model of the present study allows to observe esat-6 gene expression of M. smegmatis in HdB<Pi.
The adaptation of M. smegmatis for up to 144 h is evident in all culture media, with the exception of the mycobacteria grown in HdB<Pi (Figure 1). For which it poses instability of the bacteria to the culture, because of its continuous growth with respect to the HdB minimal medium (P>0.05) and its inactive metabolism after 144 h when changing the mycobacteria to a fresh culture, where the loss of acid-alcohol capacity is observed (Figure 2: a and b); similar results in M. tuberculosis were obtained by Rifat et al29, when limiting Pi in 7H9 medium.
This suggests, under the proposed conditions, the existence of a state of emergency, in which M. smegmatis can have different forms of adaptation, where several signal transduction pathways are involved by which mycobacteria detect and give response to the environment. These pathways can be associated with alarmone, which triggers the stringent response. An indispensable mechanism for bacterial adaptation during the transition of the exponential phase to the stationary phase30-32. In this regard, in E. coli, lack of amino acids, carbon or Pi, is followed by an increase in ppGpp levels and polyphosphate accumulation33,34.
Under limiting conditions of Pi and a sustained growth very near to HdB minimal medium (Figure 1), M. smegmatis apparently finishes the external sources and the internal energy reserves. In order to avoid this wear down of energy reserves, the stringent response has been proposed, which is characterized by ppGpp regulation that increases cyclopropanation35, inhibition and synthesis of fat acids and phospholipids36. Although the proposal of the stringent response is associated with the possibility of stabilization of the metabolism for adaptation of mycobacteria to stress conditions, that is, under conditions of abundance, polyphosphates can be used as donors of ADP, GDP and other reactions in which the Rel enzyme is involved, acting in the synthesis and hydrolysis of ppGpp5,37. However, when there are not enough Pi reserves and energy reserves are minimal, it is probable that the stringent response fails so that M. smegmatis adapts and its metabolic state does not let it respond immediately to favorable changes to renew its growth, as it happened in the present study.
An increase in ppGpp levels and polyphosphates induces the transcription of 150 genes involved in the slowdown of growth and metabolism38. Rather, what has been found in this study with respect to Pi limitation in HdB, is that growth is maintained similar to HdB minimal medium (P<0.05), where Pi is not limited. Conversely, the esat-6 gene expression in HdB<Pi that occurred at mRNA level (Figure 3B), which is comparable to 7H9 and Sauton, although the protein encoded by this gene is only secreted in Sauton medium22, it is necessary to determine whether under HdB<Pi culture conditions, esat-6 protein is secreted. This protein has also been proposed as an ATPase39, which modifies the properties of the phagosome compartment, where M. tuberculosis reside, as evidenced by aberrant retention or imperfect acquisition of a range of proteins of the host, including Rab GTPases40,41 and vacuolar (H+)-ATPase42.
In M. smegmatis and M. tuberculosis, polyphosphates regulate ppGpp synthesis through transcriptional control of relA via mprAsigE-relA pathway37,43. There are other systems of the two components that can contribute to the regulation of relA. Consequently, it is proposed as prospect to study the relationships between regulators of Pi-dependent gene expression, including SenX3-RegX3 and its relationship with genes that regulate the ESX-1 system and secretion of the proteins in M. smegmatis44. Pang et al45) confirm that in M. tuberculosis, esat-6 is not regulated by MprAB under growth in standard 7H9. Therefore, it is important to understand whether esat-6 of M. smegmatis is regulated by this system under Pi-limiting conditions. The studies of Rifat et al29, focused on Pi limitation, show that esat-6 does not express in M. tuberculosis, when they limited Pi in 7H9 medium with L-glutamate as source of organic nitrogen; however, in this model, Pi is limited and the nitrogen source is inorganic. Pang et al45 also report that at least there is an indirect regulation of ESX-1 by PhoPR. Thereon, it is suggested to research the relationships between Pi limitation and PhoPR, under the proposed model in this research.
In conclusion, M. smegmatis adapts to HdB minimal medium, HdB<C and HdB<N, and resumes its metabolic activity in fresh medium, but it does not express esat-6 gene. However, in HdB<Pi medium, M. smegmatis shows a continuous exponential growth phase, statistically similar to HdB medium (P<0.05), manifesting a metabolic state in which it loses resistant-acid capacity in the stationary phase and expresses the esat-6 gene after changing the culture medium to 96 h into the corresponding fresh culture medium. Consequently, the model of research for genetic expression under nutrient stress conditions is proposed.
Acknowledgements
Special thanks to Dr. Clara Espitia Pinzon at the Instituto Nacional de Investigaciones Biomédicas of the UNAM for the contribution of M. smegmatis strain mc2155 and to the community of the Centro de Innovación y Desarrollo Educativo and Centro de Estudios Justo Sierra, Sinaloa State.
REFERENCES
1. Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, Stoker NG. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun 2003;71(3):1134-40. [ Links ]
2. de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol 2007;189(16):6028-34. [ Links ]
3. Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2007;2(5):352-64. [ Links ]
4. Abramovitch RB, Rohde KH, Hsu FF, Russell DG. AprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol 2011;80(3):678-94. [ Links ]
5. Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA 2003;100(17):10026-31. [ Links ]
6. Siegele DA, Kolter R. Life after log. J Bacteriol 1992;174(2):345-8. [ Links ]
7. Gebhard S, Humpel A, McLellan AD, Cook GM. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 2008;154(Pt 9):2786-95. [ Links ]
8. Humpel A, Gebhard S, Cook GM, Berney M. The SigF regulon in Mycobacterium smegmatis reveals roles in adaptation to stationary phase, heat, and oxidative stress. J Bacteriol 2010;192(10):2491-2502. [ Links ]
9. Slonczewski JL, Rosen BP, Alger JR, Macnab RM. PH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci USA 1981;78(10):6271-6275. [ Links ]
10. Pepin CA, Wood HG. Polyphosphate glucokinase from Propionibacterium shermanii. Kinetics and demonstration that the mechanism involves both processive and nonprocessive type reactions. J Biol Chem 1986;261(10):4476-4480. [ Links ]
11. Lyon RH, Rogers P, Hall WH, Lichtein HC. Inducible glutamate transport in Mycobacteria and its relation to glutamate oxidation. J Bacteriol 1967;94(1):92-100. [ Links ]
12. Sritharan V, Ratledge C, Wheeler PR. Effect of homoserine on growth of Mycobacterium smegmatis: inhibition of glutamate transport by homoserine. J Gen Microbiol 1987;133(10):2781-2785. [ Links ]
13. Malhotra V, Tyagi JS, Clark-Curtiss JE. DevR-mediated adaptive response in Mycobacterium tuberculosis H37Ra: links to asparagine metabolism. Tuberculosis (Edinb) 2009;89(2):169-174. [ Links ]
14. Ganguly N, Giang PH, Gupta C, Basu SK, Siddiqui I, Salunke DM, et al. Mycobacterium tuberculosis secretory proteins CFP-10, ESAT-6 and the CFP10:ESAT6 complex inhibit lipopolysaccharide-induced NF-kappaB transactivation by down regulation of reactive oxidative species (ROS) production. Immunol Cell Biol 2008;86(1):98-106. [ Links ]
15. Welin A, Eklund D, Stendahl O, Lerm M. Human macrophages infected with a high burden of ESAT-6-expressing M. tuberculosis undergo caspase-1- and cathepsin Bindependent necrosis. PLoS One 2011;6(5):e20302. [ Links ]
16. Flint JL, Kowalski JC, Karnati PK, Derbyshire KM. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc Natl Acad Sci USA 2004;101(34):12598-12603. [ Links ]
17. GenBank of National Center for Biotechnology Information. <http://blast.ncbi.nlm.nih.gov/Blast.cgi#alnHdr_118168627>. [ Links ]
18. Cox RA, Garcia MJ. Adaptation of mycobacteria to growth conditions: a theoretical analysis of changes in gene expression revealed by microarrays. PLoS One 2013;8(4):e59883. [ Links ]
19. Ojha AK, Mukherjee TK, Chatterji D. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun 2000;68(7):4084-4091. [ Links ]
20. Berthet FX, Rasmussen PB, Rosenkrands I, Andersen P, Gicquel B. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 1998;144(Pt 11):3195-3203. [ Links ]
21. Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 2006;74(1):88-98. [ Links ]
22. Converse SE, Cox JS. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J Bacteriol 2005;187(4):1238-1245. [ Links ]
23. Lamarche MG, Wanner BL, Crepin S, Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 2008;32(3):461-473. [ Links ]
24. Smeulders MJ, Keer J, Speight RA, Williams HD. Adaptation of Mycobacterium smegmatis to stationary phase. J Bacteriol 1999;181(1):270-283. [ Links ]
25. Sanger F, Nicklen S, Chase AR. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci 1977;74(12):5463-5468. [ Links ]
26. Sambrook J, Russell DW. The condensed protocols from Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006:420-478. [ Links ]
27. Dowdy S, Weardon S, Chilko D. Statistics for research. Chapter 8: Student’s t distribution. Third ed. New Jersey: John Wiley & Sons, Inc.; 2004:178-210. [ Links ]
28. Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb) 2004;84(1-2):29-44. [ Links ]
29. Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis 2009;200(7):1126-1135. [ Links ]
30. Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell 2004;117(3):299-310. [ Links ]
31. Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol 2005;55(4):973-977. [ Links ]
32. Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3',5'-bispyrophosphate (ppGpp). J Bacteriol 2006;188(13): 4627-4634. [ Links ]
33. Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem 1990;265(20):11734-11739. [ Links ]
34. Spira B, Yagil E. The relation between ppGpp and the PHO regulon in Escherichia coli. Mol Gen Genet 1998;257(4):469-477. [ Links ]
35. Eichel J, Chang YY, Riesenberg D, Cronan JE, Jr. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigmaS). J Bacteriol 1999;181(2):572-576. [ Links ]
36. Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB). J Biol Chem 1994;269(42):26584-26590. [ Links ]
37. Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, et al. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 2007;65(2):261-276. [ Links ]
38. Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 2002;66(3):373-395. [ Links ]
39. Pallen MJ. The ESAT-6/WXG100 superfamily - and a new Gram-positive secretion system? Trends Microbiol 2002;10(5):209-212. [ Links ]
40. Via LE, Deretic D, Ulmer RJ, Hibler NS, Huber LA, Deretic V. Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J Biol Chem 1997;272(20):13326-13331. [ Links ]
41. Clemens DL, Lee BY, Horwitz MA. Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun 2000;68(5):2671-2684. [ Links ]
42. Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 1994;263(5147):678-681. [ Links ]
43. Manganelli R, Voskuil MI, Schoolnik GK, Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 2001;41(2):423-437. [ Links ]
44. Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR, Jr. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 2007;189(15):5495-5503. [ Links ]
45. Pang X, Samten B, Cao G, Wang X, Tvinnereim AR, Chen XL, et al. MprAB Regulates the espA Operon in Mycobacterium tuberculosis and Modulates ESX-1 Function and Host Cytokine Response. J Bacteriol 2013;195(1):66-75. [ Links ]
Received: May 14, 2014; Accepted: September 09, 2014











 texto en
texto en 


