Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista mexicana de ciencias agrícolas
versión impresa ISSN 2007-0934
Rev. Mex. Cienc. Agríc vol.12 no.3 Texcoco abr./may. 2021 Epub 02-Mayo-2022
https://doi.org/10.29312/remexca.v12i3.2619
Articles
Phytophthora species associated with irrigation water in the Culiacán Valley
1Facultad de Agronomía-Universidad Autónoma de Sinaloa. Carretera Culiacán-Eldorado km 17.5, Culiacán Rosales, Sinaloa, México. AP. 25. CP. 80000. (moisesyj@uas.edu.mx; clopezorona@uas.edu.mx; tafoya@uas.edu.mx; alfonsolopezurquidez@uas.edu.mx).
2Facultad de Química-Universidad Autónoma de Querétaro. Centro Universitario Hidalgo s/n, Cerro de las Campanas, Querétaro. CP. 76010. (ser69rom@gmail.com).
In Mexico there are few studies describing aquatic bodies used for agriculture and the role of irrigation water as a source of inoculum, persistence and dispersal of phytopathogenic parasites, such as oomycetes that inhabit aquatic environments with the capacity to infect a wide range of cultivated hosts. Due to the lack of information on which oomycetes genus and species are present in the water, their identification is of paramount importance in managing alternative management and control of them for agricultural production in the region. In order to determine which oomycetes are isolated in surface waters used for irrigation in the Culiacán Valley, Sinaloa, in the period from September 2018 to January 2019, in different agricultural water networks (reservoir, ponds, rivers and canals), floating traps made with polypropylene bags and two pear fruits that were placed that served as bait. Oomycete isolates were obtained, which were identified based on their morphological characteristics and DNA sequences (with ITS 4/6, COX and NADH initiators), pathogenicity tests were also performed on plants and fruits of the solanacea and cucurbitaceae families. The organisms were isolated and identified: Phytophthora parsiana, P. virginiana, P. lagoariana, P. capsici and P. hydropathica. All Phytophthora species were pathogenic, they had the ability to infect plants and fruits, causing symptoms of wilting and rotting in the inoculated vegetables.
Keywords: nucleic acids; oomycetes; pseudo-fungus; vegetables
En México existen escasos estudios que describen a los cuerpos acuáticos empleados para la agricultura y el rol que tiene el agua de riego como fuente de inóculo, persistencia y dispersión de parásitos fitopatógenos, tal es el caso de los oomycetes que habitan ambientes acuáticos con capacidad de infectar un amplio rango de hospedantes cultivados. Debido a la falta de información sobre qué géneros y especies de oomycetes están presentes en el agua, su identificación es de suma importancia para gestionar alternativas de manejo y control de estos para la producción agrícola en la región. Con el objetivo de determinar qué oomycetes se aíslan en aguas superficiales empleadas para la irrigación en el Valle de Culiacán, Sinaloa, en el periodo comprendido de septiembre de 2018 a enero de 2019, en diferentes redes hídricas agrícolas (presas, estanques, ríos y canales), se colocaron trampas flotantes elaboradas con bolsas de polipropileno y dos frutos de pera que sirvieron como cebo. Se obtuvieron aislados de oomycetes, los cuales se identificaron en base a sus características morfológicas y secuencias de ADN (con los iniciadores ITS 4/6, COX y NADH), también se realizaron pruebas de patogenicidad en plantas y frutos de las familias de solanáceas y cucurbitáceas. Se aislaron e identificaron los organismos: Phytophthora parsiana, P. virginiana, P. lagoariana, P. capsici y P. hydropathica. Todas las especies de Phytophthora resultaron patogénicas, tuvieron capacidad de infectar plantas y frutos, causando síntomas de marchitez y pudrición en las hortalizas inoculadas.
Palabras clave: ácidos nucleicos; hortalizas; oomycetes; pseudohongo
Introduction
The organisms of the genus Phytophthora, belong to the kingdom Cromista or Stramenopila, are inhabitants of aquatic and terrestrial environments, with high capacity for reproduction, dissemination and survival, particularities that facilitate their establishment in agricultural or forestry fields (Erwin and Ribeiro, 1996; Chen et al., 2017, Hon, 2018; Judelson et al., 2019). This genus includes species with a wide range of hosts such as P. cinnamomi which is capable of parasitizing around 5 000 species of plants (Hardham et al., 2018) and P. capsici, known for causing economic losses in solanáceae and cucurbitaceae plantations grown around the world (Vega et al., 2017).
About 30 species of Phytophthora were collected in bodies of water such as ponds, rivers, canals, reservoirs and hydroponic crops (Aram and Curl, 2018) in addition, the genus Phytophthora, like other phytopathogenic oomycetes, strictly requires water for reproduction which begins with the formation of sporangiophores and sporangia, within which zoospores are formed for further release and dissemination; these zoospores, are able to infect susceptible plants and start the cycle of a disease (Kang et al., 2017).
Zoospores are the main infective propelling spread by free water, so it is possible to recover them regularly from surface water (Hon, 2018). Examples of isolated water species are often: P. cactorum, P. parasitica, P. citrícola, P. gonapodyides, P. cambivora, P. hydropathica and P. drechsleri, among others (Loyd, 2014; Redekar and Park, 2018; Ristvey et al., 2019). In Mexico, there are about 17 species belonging to the genus Phytophthora reported as parasites of plants of agricultural importance, the most common are: P. capsici, P. infestans and P. cinnamomi; however, information on the attribution of surface water as a source of inoculum and dispersal of organisms of the genus Phytophthora is scarce.
Thus, Álvarez et al. (2016) were able to isolate and identify the species P. hydropathica and P. drechsleri from different water channels for agricultural use in Culiacán, Sinaloa, Mexico. On the other hand, the identification of oomycete genus and species was commonly based on their morphological and cultural characteristics; however, erroneous identifications were common, because some species share these characteristics; currently, these practices are complemented by biotechnological techniques based on the extraction and sequencing of nucleic acids, which allows the identification of organisms effectively and concretely (Trzewik et al., 2016). Therefore, the objective of this study was to determine the incidence of species of the genus Phytophthora in surface waters for agricultural use in Culiacán, Sinaloa and to determine their pathogenic potential in plants grown in Mexico.
Materials and methods
Obtaining, purifying and conservation of isolates Phytophthora spp.
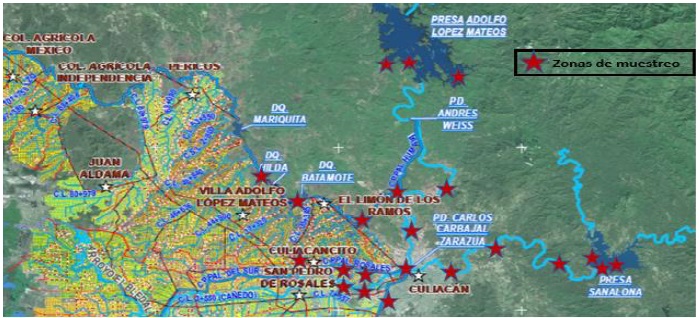
Using a map of the location of dams, dikes, ponds, rivers and water storage and distribution of the water used in agricultural activities, of the area of influence of irrigation district number 010 of the Culiacán Valley, Sinaloa, granted by the National Water Commission (CONAGUA) (Figure 1), 37 sampling sites were randomly selected in 28 locations (Table 1).
Table 1 Geographic location of the sampling sites to obtain isolates of Phytophthora spp., Culiacán, Sinaloa, 2018-2019.
Locality |
Coordinates of the sampling site |
Isolation |
Adolfo López Mateos Prey (The Varejonal) |
25°10'08'' N 107°26'10'' W 25°09'10'' N107°26'11'' W 25°09'56'' N 107°26'30'' W |
PV1, PV2, PV3, PV4, PV5, PV6, PV7, PV8, PV9, PV10, PV11, PV12 |
La Presita |
24°55'49'' N 107°25'39'' W |
PR13, PR14, PR15, PR16 |
Empaque Castro |
24°55'28'' N 107°31'59'' W |
EC17, EC18, EC19, EC20 |
The Tamarindo |
24°55'07'' N 107°33'32'' W 24°55'11'' N 107°32'57'' W |
TR21, TR22, TR23, TR24 |
Empaque Valle del sol |
24°53'20'' N 107°29'17'' W |
EVS25, EVS26, EVS27, EVS28 |
Sanalona Prey |
24°48'37'' N 107°08'11'' W 24°48'46'' N 107°08'14'' W 24°48'49'' N 107°08'30'' W |
PS33, PS34, PS35, PS36, PS37, PS38, PS39, PS40, PS41, PS2, PS43 |
Tamazula River |
24°49'00'' N 107°11'16'' W 24°48'53'' N 107°21'29'' W |
RT44. RT45, RT46, RT47, RT48, RT49 |
Humaya River |
24°51'48'' N 107°24'25'' W |
RH50, RH51, RH52, RH53, RH54, RH55 |
Botanical garden |
24°49'23'' N 107°23'06'' W 24°49'24'' N 107°23'04'' W |
JB56, JB57, JB58, JB59, JB60, JB61 |
Imala |
24°51'21'' N 107°13'11'' W |
IMA62, IMA63, IMA64, IMA65 |
Three Rivers |
24°48'43'' N 107°24'24'' W 24°48'24'' N 107°24'33'' W |
3R66, 3R67, 3R68, 3R69, 3R70, 3R71 |
Bellavista |
24°48'45'' N 107°27'56'' W |
BELL74 and BELL75 |
Bachigualato |
24°46'59'' N 107°26'48'' W |
BACH76 and BACH77 |
Aguaruto |
24°47'49'' N 107°30'24'' W |
AGUA78 and AGUA79 |
Batan Field |
24°46'10'' N 107°30'19'' W 24°45'40'' N 107°30'45'' W |
CB80 and CB81 |
Moroleón Field |
24°41'14'' N 107°31'09'' W |
CM82 and CM83 |
Cardenal Field |
24°45'10'' N 107°28'39'' W |
CC84 and CC85 |
Primavera Dam |
24°43'48'' N 107°24'10'' W |
DP86 and DP87 |
Hazera Station |
24°43'32'' N 107°27'24'' W |
HAZ88 and HAZ89 |
Enza Zaden Station |
24°40'58'' N 107°28'47'' W |
EZ90 and EZ91 |
Monsanto Station |
24°40'37'' N 107°28'50'' W |
MON92 and MON93 |
Divemex Field |
24°39'46'' N 107°26'58'' W |
DIV94 and DIV95 |
US Agriseeds Station |
24°38'11'' N 107°26'38'' W |
US96 and US97 |
Syngenta Station |
24°37'08'' N 107°26'41'' W |
SYN98 and SYN99 |
Rijk Zwaan Station |
24°36'12'' N 107°26'41'' W |
RZ100 and RZ101 |
Bayer Station |
24°36'14'' N 107°27'22'' W |
BAY102 and BAY103 |
Costa Rica Dam |
24°35'44'' N 107°18'33'' W |
CR104 and CR105 |
Faculty of Agronomy |
24°37'28'' N 107°26'34'' W |
PP106 and PC107 |
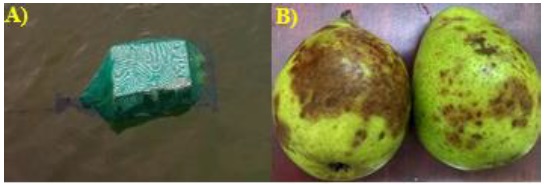
The sampling period from September 2018 to January 2019, by sampling site was placed a trap made of polypropylene net bag, closed from the front with a rope of the same material, together, that rope was used to fix the trap at some point in the slope (Figure 2).
These traps, inside, contained two injury-free pear fruits, washed with simple water and superficially disinfested with 96° ethanol (Soto et al., 2017). The traps remained suspended for 48 hours, then collected and transported to the phytopathology laboratory of the Faculty of Agronomy of the Autonomous University of Sinaloa. The fruits with obvious lesions were washed with sterile distilled water, dried with sterile brown paper and disinfected with 96° degree ethanol.
The insulations were made from cuts of 1 cm2 taken from the brown lesions on the fruits (Figure 2) and every five fragments were placed equidistantly in petri boxes containing selective culture medium made with corn-agar flour (HMA, 17 g L-1), penicillin (10 µg L-1), ampicillin (200 µg L-1), rifampicin (10 µg L-1), pentachloronitrobenzene (PCNB, 25 µg L-1) (PARP) (Jeffers, 2006; Soto et al., 2017).
Petri boxes were left two days at room temperature (26 ±2 °C). In order to purify and conserve individual strains from the developed colonies of the sick tissue, it was transferred to HMA culture medium using the hypha tip technique, placed 48 h at room temperature, once the colonies were developed, with tweezers, five pieces of 0.5 cm diameter of culture medium with mycelial growth were taken individually from each isolation to later place them in microvial tubes (5 ml) containing sterile distilled water and were stored at 15 °C for further studies (Jeffers, 2006; Abbot et al., 2012; Soto et al., 2017).
Morphological and cultural characteristics
To perform morphological characterization of organisms, of the strains preserved in microvial tubes, fragments of mycelium were individually taken which were sown in the culture medium containing distilled water (800 ml), V8 juice (200 ml), CaCO3 (2 g) and agar (15 g) (V8A), in order to induce the growth and reproduction of the pathogen, petri boxes remained 72 h at room temperature, subsequently, with tweezers, five 0.5 cm diameter fragments were taken from each insulation and placed in Petri boxes of 90 mm in diameter containing distilled water and aqueous soil extract that served for induction into the formation of sporangia.
The boxes were incubated at room temperature for 72 h (Martin et al., 2004; Jeffers, 2006; Soto et al., 2017). In order to induce other morphological structures such as: chlamydospores, hyphal swellings, antheridia, oogonia and oospores, of the strains preserved in microvial tubes were individually taken fragments of mycelium that were sown in Petri boxes containing V8Aclarified culture medium (V8 juice subjected to 4 000 RPM for 20 min, V8AC) and placed for 21 days under room temperature and dark conditions.
The type of reproductive sexual compatibility was determined, for which each isolation was confronted with reference lineages of P. capsici and P. drechsleri with affinity A1/A2 (organisms provided from the strain of phytopathogenic fungi of the Autonomous University of the West Campus Los Mochis, Sinaloa). All the conserved strains were confronted in V8AC culture medium, for which a cylinder of 0.5 cm diameter of culture medium with mycelial growth was planted at one end of the box and at the opposite end a cylinder of the strain A1 or A2 was placed individually, and then keep them at room temperature and darkness for twenty-one days (Jeffers, 2006; Abbot et al., 2012; Soto et al., 2017). The morphological characterization of insulation was performed by observing the structures developed with optical microscope (Leica DM100 with eye meter) and comparing with a pictorial key of species of the genus Phytophthora (Abad et al., 2012).
Molecular identification
DNA extraction
The Method of Cetyltrimethylammonium Bromide (CTAB), which consists of a series of steps presented below, was used for DNA extraction: 100-200 mg of mycelium developed on medium dextrose potato agar (39 g L-1, PDA) were collected. The mycelium was placed on individual mortars, liquid nitrogen was added to each sample and crushed with a pistil until a fine powder was obtained.
The powder was individually transferred to Eppendorf tubes of 2 ml held in freezing and 1 ml of extraction buffer was added (CTAB 2%, TRISH-HCl 100 mM PH 8, EDTA 20 mM PH 8, NaCl 1.4 M, β-mercaptoethanol 2%). The tubes were placed in vortex for a minute. The tubes were inserted in a centrifuge at 12 000 RPM for 15 minutes. The over natant was recovered and transferred individually to new Eppendorf tubes of 2 ml containing 1 ml of chloroform-isoamyl alcohol solution (49:1), the contents of the tubes were mixed with the recovered phase.
Samples were placed in vortex for 10 min. A second aqueous phase recovery and a second transfer to new Eppendorf tubes were performed. Sodium acetate 3M was added with 1 ml isopropyl alcohol with value 1/10 of the recovered volume and the tubes were stored at 20 °C by 20 min (tubes can be stored for up to 24 hours to maximize DNA production). The tubes were centrifuged at 12 000 RPM for 15 min, then they were decanted without wasting the precipitated DNA content. 1 ml of ethanol was added individually and samples were centrifuged for 3 min. The over natant was extracted cautiously without touching the solid phase, the tubes with the solid phase were placed at room temperature for evaporation of liquids.
DNA was dissolved in 50 µl of buffer TE. DNA characteristics were calculated with a Nanodrop at 260 nM, the quality was determined with the ratio 280/260, the integrity was visualized in a 0.8% agar gel electrophoresis chamber with 1X TAE buffer.
DNA amplification, purification and sequencing
The reaction mixture consisted of 1 µl of DNA (50 ng µl), 0.5 µl of dntp’s, 0.25 Taq DNA polymerase, 2.5 µl buffer, 0.625 µl of each first, 20.175 µl of water in a total reaction volume of 25 µl. DNA amplification was performed on a thermocycler (Bio rad T100). The thermocycler sequence was as follows: an initial denaturation at 94 °C by 2 min; 35 denaturation cycles at 94 °C by 30 s (60 s for COX and NADH), alignment by 30 s and extension to 72 °C by 60 s, the final extension at 72 °C per 10 min. The alignment temperature was 52 °C for COX, 53 °C for NADH and 62 °C for ITS (Table 2), whose successful amplification was confirmed by the electrophoresis gel.
Table 2 Initiators used for DNA amplification of Phytophthora spp. (Kroon et al., 2004).
First |
First sequence |
Size (pb) |
ITS4C0569ITS6C0570 |
TCCTCCGCTTATTGATATGCGAAGGTGAAGTCGTAACAAGG |
930 |
COXF4NCOXR4N |
GTATTTCTTCTTTATTAGGTGCGTGAAGTAATGTTACATATAC |
972 |
NADHF1NADHR1 |
CTGTGGCTTATTTTACTTTAGCAGCAGTATACAAAAAGCAAC |
897 |
The PCR products were purified using the SV Gel and PCR CleanUp System (Promega, USA) wizard kit. Both chains of the amplicon were sequenced at the Macrogen, Korea facility. The results of the isolations were analyzed by Blast comparing sequences of isolates deposited in the specialized database Phytophthora (http://www.phytophthoradb.org, http://www.phytophthora-ID.org) and NCBI (http://www.NCBI.com) to determine the percentage of similarity.
Pathogenicity tests
Obtaining seedlings
In order to verify the pathogenicity of each of the isolates, saladette tomato seedlings (cv SV-3543) were produced, bell chili (cv Caravaggio) and slicer cucumber (cv Luxell). The seeds of each species were individually sown in polystyrene trays with 256 cavities filled with wet peat moss substrate (Berger), once sown, the trays remained in greenhouse conditions.
Seven days after the emergence of seedlings, they were transplanted into plastic pots containing 2 kg of peat moss substrate and they watered every three days with a mixture of water and Maxigrow (percentage composition g L-1: organic extracts 112, auxins 0.09, gibberellins 0.1, cytokinin 1.5, nitrogen 6.6, phosphorus 13.3, potassium 13.3, calcium 2, magnesium 4, iron 17.2, zinc 26.5, manganese 13.3 and copper 13.3).
Pathogenicity tests on plants
For the inoculation of seedlings each insulation was transferred to Petri boxes with medium PARP and they were incubated for 10 days, subsequently, 1 cm2 cuts of culture medium with mycelial growth were taken from each insulation and placed individually on the stems of the plants flush with the substrate (25 days after transplantation), immediately the inoculum was covered with substrate; five days after inoculation, the irrigations were more abundant to promote infection. The pathogenicity of the insulation was determined by daily observation over a period of 20 days.
Pathogenicity tests on fruits
Tomato, chili and cucumber fruits with physiological maturity (free of superficial lesions), washed with sterile distilled water and disinfested with 70% ethanol, were inoculated and placed inside wet chambers in plastic containers, inoculated with cuts of 1 cm2 of Parp culture medium with 10 days of growth and sterile distilled water was added to create a favorable environment for the organism.
Three days after inoculation, the severity of the disease was determined based on the measurement of the surface damaged in cm2 by the pathogen. Four repetitions and their respective control (without inoculation of the pathogen) were used in the pathogenicity tests of plants and fruits. Re-isolations were made from sick tissue taken from inoculated plants to verify the postulates of Koch.
Results and discussion
17 isolates belonging to the genus Phytophthora were obtained, which were classified into two groups taking into account their morphological similarity (Table 3): the first group consisted of 14 isolations that developed simple sporangiophores, non-papillate ovoid sporangia with average length and width dimensions 47.5-60 x 26.25-32.5 µm (TR21, PS36, 3R68, CR104 and AGUA79), 47.5-57.5 x 26.25-32.5 µm (PV1, PV11, JB61, CC84 and CC85), 45-60 x 22.5-32.5 µm (RT45 and RT47) and 45-57.5 x 25-32.5 µm (JB61 and 3R70), with nested and extended proliferation, simple sporangiophores and irregular swellings present (Figure 3). The type of compatibility of these insulations with A1 and A2 affinity strains of P. drechsleri and P. capsici could not be identified.
Table3 Phenotypiccharacteristics of Phytophthora spp. insulation.
Group |
Isolation |
Form of sporangia |
Proliferation of sporangia |
Mycelial swelling |
Dimensions (length and width) of the sporangium (μm) |
Compatibility |
Oospores |
1 |
TR21, PS36, 3R68, CR104 and AGUA79 |
Non-papillated ovoid |
Nested and extended |
Present |
47.5-60 x 26.25-32.5 |
Stranger |
- |
PV1, PV11, JB61, CC84 and CC85 |
Non-papillated ovoid |
Nested and extended |
Present |
47.5-57.5 x 26.25-32.5 |
Stranger |
- |
|
RT45 and RT47 |
Non-papillated ovoid
|
Nested and extended |
Present |
45-60 x 22.5-32.5 |
Stranger |
- |
|
JB61 and 3R70 |
Non-papillated ovoid |
Nested and extended |
Present |
45-57.5 x 25-32.5 |
Stranger |
- |
|
2 |
PC107, MJB72 and MPV31 |
Ovoid papillated |
Simple |
Absent |
40-68 x 17-30 |
A1 |
Plerotica without ornaments (22-31 µm) |

Figure 3 Sexual and asexual structures of oomycetes. A-D) ovoid sporangia Phytophthora spp.; E) papillated sporangia of P. capsici (isolation PC107); F) release of zoospores (isolation TR21); G) nested proliferation (PV11 isolation); H) extended proliferation (isolation JB61); I) swelling of the intercalary mycelium (isolation RT45) and J) oospore of P. capsici (isolation MPV31).
The second group consisting of three isolations (PC107, MJB72 and MPV31), those who developed papillate ovoid sporangia, long and wide average dimensions of 40-68 x 17-30 µm, simple sporangiophores, simple sporangia (without proliferation) and papillates; without the presence of swellings in the hyphae. Sexual reproduction with A1 affinity of P. capsici, with formation of pleroticas oospores, without ornaments and average diameter of 22-31 µm (Figure 3).
Comparison of these characteristics with those described in the taxonomic keys of Erwin and Ribeiro (1996) and Abad et al. (2012), coincide with the species Phytophthora parsiana, P. virginiana, P. lagoariana, P. hydropathica and P. capsici. PCR reactions with ITS initiators (4-6) amplified a product approximately 941 bp, with COX 972 pb initiators and NADH 897 pb initiators.
The consensus sequences showed results with percentages of similarity to Phytophthora virginiana: 99.42 (isolation PV1), 99.47 (isolation PV11), 99.52 (isolation JB61), 99.60 (isolation CC84) and 99.78 (isolation CC85), for Phytophthora parsiana: 99 (isolation RT45) and 99.22 (isolation RT47), for Phytophthora lagoariana: 98 (isolated JB61) and 98.52 (isolated 3R70), for Phytophthora hydropathica. 97 (isolation TR21), 98 (isolation PS36), 98.5 (isolation 3R68), 99 (isolation CR104) and 99.64 (isolation AGUA79) and for Phytophthora capsici of 99.48 (isolation PC107), 99.70 (isolation MJB72 and 100% (isolation MPV31) (Table 4).
Table 4 Phytophthora species identified by isolation.
Isolates |
Genus |
Access |
Similarity (%) |
PV1, PV11, JB61, CC84 and CC85 |
Phytophthora virginiana |
MT232849 |
99.42-99.78 |
RT45 and RT47 |
Phytophthora parsiana |
MT232850 |
99-99.22 |
JB61 and 3R70 |
Phytophthora lagoariana |
MT232839 |
98-98.52 |
TR21, PS36, 3R68, CR104 and AGUA79 |
Phytophthora hydropathica |
MT339042 |
97-99.64 |
PC107, MJB72 and MPV31 |
Phytophthora capsici |
MT232875 |
99.48-100 |
The species identified in this research are regularly found in surface water (Zappia et al., 2014, Yang and Hong, 2015). The 17 isolations obtained were pathogenic, because after inoculation they all caused symptoms of seedling withering and root necrosis; in fruits caused symptoms of rot (Figure 4).

Figure 4 Pathogenicity tests of the species P. hydropathica. A-C) tomato, chili and cucumber plants, left inoculated plants and right witness plants. D-F) stems and roots of tomato, chili and cucumber plants, left inoculated plants and right witness plants. G-I) inoculated fruits of tomato, chilli and cucumber.
The isolations of P. capsici originated withering plants three days after inoculation, while those same symptoms appeared 10 days after inoculation with the rest of the species. These results corroborate the pathogenicity report of Phytophthora parsiana, P. virginiana, P. lagoariana, P. hydropathica and P. capsici on tomato, chili and cucumber plants described by Cline (2008); Lamour et al. (2012); Álvarez et al. (2016).
This research verifies the existence of unreported phytopathogenic species that represent a risk to the health of horticultural crops in the region. The extraction, amplification and sequencing of regions of the DNA of organisms using the molecular techniques used in this research (Its, Cox and Nadh), allowed their identification when determining homology levels between 97 and 100% in the NCBI database and with that, greater efficiency than with the traditional identification morphological characterization.
This surface water study demonstrates the presence in that habitat of Phytophthora virginiana, Phytophthora parsiana, Phytophthora lagoariana, Phytophthora hydropathica and Phytophthora capsici; also, the importance of water as an inoculum source and dispersion of these pathogens. In turn, it expands the range of studies for future research on the survival of such pathogens, discovering new natural hosts in the area, determining other possible means of dissemination, finding new hosts of economic importance, identifying whether there is the presence of other types of pathogens of agricultural importance in water, and in this way consider the potential risk that this type of organisms represents for the agriculture in the region.
Conclusions
In surface waters for agricultural use of irrigation district number 10 of the Culiacán Valley, Sinaloa, 17 isolations were collected that corresponded to the organisms P. virginiana, P. parsiana, P. lagoariana and Phytophthora capsici, all with the capacity to infect plants and fruits of tomato, chili and cucumber.
Acknowledgments
To CONACYT for the financing the studies of Josué Cárdenas Rodríguez and the Autonomous University of Sinaloa-Faculty of Agronomy for supporting this research.
REFERENCES
Abad, Z. G. and Balci, Y. 2012. Identification and detection of Phytophthora: reviewing our progress, identifying our needs. United States of America. Plant Dis. 96(8):1081-1103. Doi: 10.1094/PDIS-12-11-1036-FE. [ Links ]
Álvarez, B; García, R. S; Valdez, J. B.; León, J; Allende, R. and Fernández, S. P. 2016. Phytophthora hydropathica and Phytophthora drechsleri isolated from irrigation channels in the Culiacan Valley. México. Rev. Mex. Fitopatol. 35(1):20-39. Doi: 10.18781/R.MEX.FIT.1606-1. [ Links ]
Aram, K. and Rizzo, D. M. 2018. Distinct trophic specializations affect how to Phytophthora ramorum and clade 6 phytophthora spp. colonize and persit on Umbellularia californica leaves in streams. California. Phytopatlogy. 108(7):858-869. Doi: 10.1094/phyto-06-17-0196-R. [ Links ]
Chen, X. and Wang, Y. 2017. Phytophthora sojae. In: biological invasions and its management in China. Simberloff, D. (Ed). Tennessee, USA. 252 p. [ Links ]
Cline, E.; Farr, D. F. and Rossman, A. Y. 2008. A Synopsis of Phytophthora with accurate scientific names, host range, and geographic distribution. Baltimore. Plant Health Progress. 9(1):32-43. Doi: 10.1094/PHP-2008-0318-01-RS. [ Links ]
Erwin, D. C. and Ribeiro, O. K. 1996. Phytophthora diseases worldwide. Minnesota. The American Phytopathological Society. 562 p. [ Links ]
Hardham, A. and Blackman, L. M. 2018. Pathogen profile update Phytophthora cinnamomi. Australia. Plant Pathol. 19(2):260-285. Doi: 10.1111/mpp.12568. [ Links ]
Hon, H. O. 2018. The taxonomy and biology of Phytophthora and Pythium. New York. J. Bacteriol. Mycol. 6(1):40-45. Doi:10.15406/jbmoa.2018.06.00174. [ Links ]
Jeffers, S. N. 2006. Identifying species of Phytophthora. Clemson. Plant Dis. 83(12):1129-1136. [ Links ]
Judelson, H. S. and Ah-Fong, A. M. 2019. Exchanges at the plant-oomycete interface that influence disease. Plant Physiol. 179(4):1198-1211. Doi: 10.1104/pp.18.00979. [ Links ]
Kang, D. S; Min, K. J; Kwakl, A. M; Lee, S. Y. and Kang, H. W. 2017. Defense response and suppression of Phytophthora blight disease of pepper by water extract from spent mushroom substrate of Lentinula edodes. Korea. Plant Pathol. J. 33(3):264-275. Doi: 10.5423/PPJ.OA.02.2017.0030. [ Links ]
Kroon, L. P.; Bakker, F. T; Van, G. B.; Bonants, P. M. and Flier, W. G. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol. 41(8):766-782. Doi: 10.1016/j.fgb.2004.03.007. [ Links ]
Lamour, K. H.; Stam, R.; Jupe, J. and Huitema, E. 2012. The oomycete broad-host-range pathogen Phytophthora capsici. United States. Mol. Plant Pathol. 13(4):329-337. Doi.org/10.1111/j.1364-3703.2011.00754.x. [ Links ]
Loyd, A. L.; Benson, D. M. and Ivors, K. L. 2014. Phytophthora populations in nursery irrigation water in relationship to pathogenicity and infection frequency of Rhododendron and Pieris. North Caroline. Plant Dis. 98(9):1213-1220. Doi: 10.1094/PDIS-11-13-1157-RE. [ Links ]
Martin, F. N.; Tooley, P. W.; and Blomquist, C. 2004. Molecular detection of Phytophthora ramorum, the causal agent of sudden oak death in California, and two additional species commonly recovered from diseased plant material. Phytopathology. 94(6):621-31. Doi: 10.1094/phyto.2004.94.6.621. PMID, 18943487. [ Links ]
Redekar, N. M.; Eberhart, J. L. and Parke, J. L. 2018. Diversity of Phytophthora, Pythium and Phytopythium species in recycled irrigation water in a container nursery. Oregon. Phytobiomes J. 3(1):31-45. Doi: 10.1094/pbiomes-10-18-0043-r. [ Links ]
Ristvey, A. G.; Belayneh, B. E. and Lea, J. 2019. A comparison of irrigation-water containment methods and management strategies between two ornamental production systems to minimize water security threats. United States of America. J. Water. 11(12):2558. Doi: 10.3390/w11122558. [ Links ]
Soto, A.; Rodríguez, G.; Fernández, Y. L.; Pedraza, M. E.; López, L.; Díaz, M. y Fernández, S. P. 2017. Protocolos de aislamiento y diagnóstico de Phytophthora spp. enfoque aplicado a la investigación. México. Rev. Mex. Cienc. Agríc. 8(8):1867-1880. [ Links ]
Trzewik, A.; Katarzyna, N. and Orlikowska, T. 2016. A simple method for extracting DNA from rhododendron plants infected with Phytophthora spp. for use in PCR. Polonia. J. Plant Protec. Res. 56(1):96-100. Doi: 10.1515/jppr-2016-0014. [ Links ]
Vega, J. C.; Beltrán, H. S.; Sevillano, J. and Moffet, P. 2017. Non-host plant resistance against Phytophthora capsici is mediated in party by members of the I2 R gene family in Nicotiana spp. Canada. Frontiers in Plance Sci. 8(1):205. Doi:10.3389/fpls.2017.00205. [ Links ]
Yang, X. and Hong, C. X. 2015. Diversity and populations of Phytophthora, Phytopythium and Pythium species recovered from sediments in an agricultural run-off sedimentation reservoir. Virginia. Plant Pathol. 65(1):1118-1125. Doi: 10.1111/ppa.12488. [ Links ]
Zappia, R. E.; Huberli, G. E.; Hardy, J. and Bayliss, K. L. 2014. Fungi and oomycetes in open irrigation systems: knowledge gaps and biosecurity implications. Australia. Plant Pathol. 63(5):961-972. Doi: 10.1111/ppa.12223. [ Links ]
Received: December 01, 2020; Accepted: March 01, 2021











 texto en
texto en