Introduction
Nicaragua has wide coasts along the Pacific Ocean (410 km) and the Caribbean Sea (530 km) that are used for fishing and aquaculture, representing a significant contribution to the national economy (ProNicaragua 2019). Fishing in Nicaragua not only contributes to the food security of the population by increasing food supplies, providing animal protein and other nutrients, but also by generating jobs and income to meet other needs (FAO 2014). Small-scale fishing activities on the Pacific coast of the country have targeted mainly snappers (Lutjanus spp.), groupers (Epinephelus striatus), mullets (Mugil spp.), pomfret (Brama brama), and croakers (Cynoscion spp.) (FAO 2018). However, the intensification of fishing activities has been accompanied by an increase in emerging and reemerging diseases (Ghaly et al. 2010).
Lutjanus guttatus and Mugil cephalus fishes caught in the wild can carry pathogenic bacteria, such as Aeromonas spp., Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus (Gomez-Gil et al. 2007, Hassen et al. 2020), whereby they could be considered a reservoir or vehicle for foodborne infections and, therefore, a threat to public health (Bonnin-Jusserand et al. 2019). Furthermore, fishes caught in shallow waters adjacent to coastal areas, where marine and fresh waters converge, may be infected by bacteria from human sources, such as Salmonella spp., Listeria monocytogenes, and Staphylococcus aureus (Feliatra et al. 2020).
Bacteria of the genus Staphylococcus can be transmitted by fish, but unlike the genus Vibrio, they are not typical of marine environments and are thus not considered autochthonous (Romero-Jarero and Negrete-Redondo 2011). In addition, Vibrio species such as V. cholerae and V. parahaemolyticus, which are present in aquatic ecosystems, are recognized for causing gastrointestinal tract infections in humans with high resistance to antibiotics (Bonnin-Jusserand et al. 2019). Other bacteria have also been identified in fish, such as Escherichia coli, Enterobacter cloacae, Citrobacter spp., Salmonella Paratyphi A and B, Salmonella Enteritidis, Clostridium botulinum, Enterobacter spp., Serratia ssp., Enterococcus faecalis, and Staphylococcus epidermidis (Romero-Jarero and Negrete-Redondo 2011).
In Nicaragua, there are no microbiological studies on marine fishes. Therefore, the aim of this study was to identify Vibrio spp. and other pathogenic bacteria in fishes from 4 communities on the Pacific coast of Nicaragua. This is the first study describing the frequency of bacteria in fishes landed by artisanal fisheries in Nicaragua.
Materials and methods
Sampling
The study was carried out in 4 fishing communities (El Tránsito: 12º04′24′′ N, 86º72′21′′ W; Las Peñitas: 12º34′28′′ N, 87º02′38′′ W; Poneloya: 12º37′10′′ N, 87º06′00′′ W, Jiquilillo: 12º72′48′′ N, 87º45′86′′ W) on the western Pacific coast of Nicaragua, between October and November 2019. A total of 62 samples were analyzed and they were distributed in proportion to the number of fishes caught per community: 31 fishes from the Jiquilillo community, 13 fishes from El Tránsito, 10 fishes from Las Peñitas, and 8 fishes from Poneloya (Fig. 1). The fish species analyzed were Lutjanus colorado (n = 18), Caranx latus (n = 14), M. cephalus (n = 13), B. brama (n = 9), and Pomadasys spp. (n = 8).
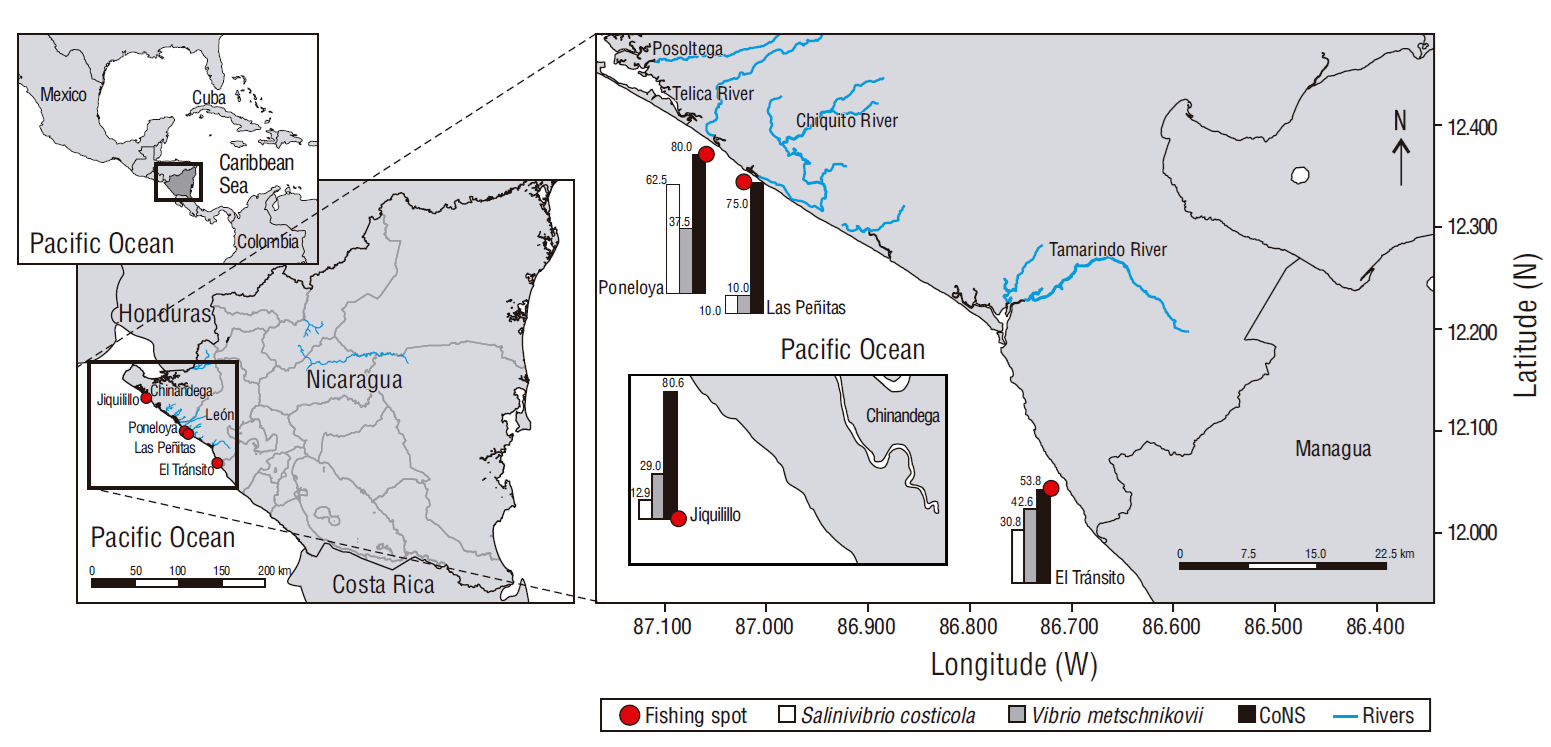
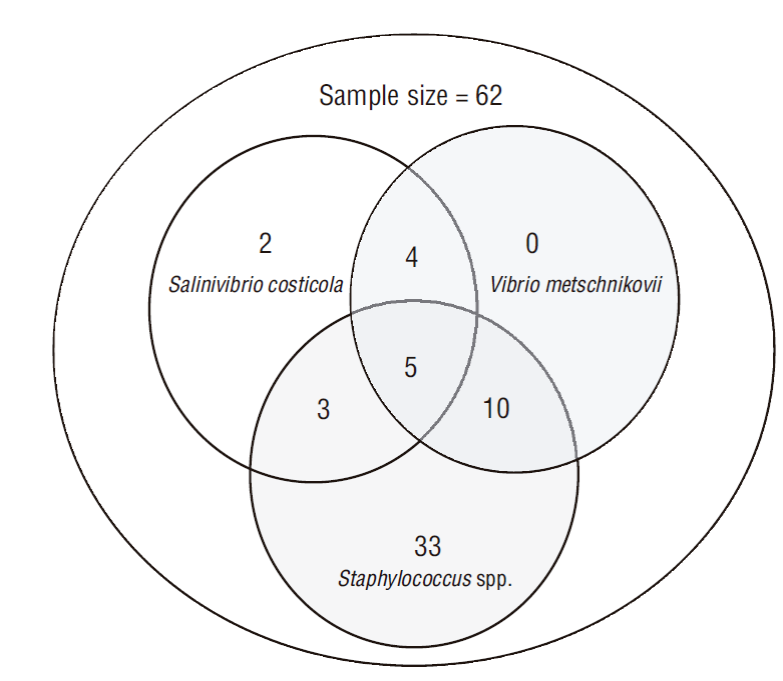
Figure 1 Relative frequency of bacteria isolated from fishes caught in the Pacific Ocean off Nicaragua. CoNS, coagulase-negative staphylococci.
An external assessment of the fishes was made for macroscopic lesions such as bleeding in the skin, fins, and gills; ulcers; blisters; and peeling. Relevant variables were recorded: fish species, capture time (data provided by the fisherman), origin (coordinates), and weight. The samples were individually introduced in sterile bags without any additive and were transported in a portable cooler with refrigerant packs at temperatures between 4 and 8 ºC for immediate processing (less than 1 h) in the Laboratorio de Microbiología Veterinaria at the Centro Veterinario de Diagnóstico e Investigación, Escuela de Ciencias Agrarias y Veterinarias, Universidad Nacional Autónoma de Nicaragua, León.
Bacteriological analysis
Determination of Vibrio load
Fish surfaces were cleaned with 95% alcohol, and the necropsy was performed using a sterile scalpel. A 1-g portion of the dorsal muscle was weighed aseptically; it was homogenized with 9 mL of sterile peptone water (SPW) using a mortar (previously sterilized at 160 ºC for 1 h) and a vortex mixer (Fisherbrand; Waltham, MA, USA) for 2 min. Immediately afterwards, serial tenfold dilutions were made from the first mixture, adding 9 mL of SPW plus 1 mL of the previous mixture, in such a way that the dilutions 1:10, 1:100, 1:1,000 were obtained. Then, 100 µL of each dilution were plated on thiosulphate-citrate-bile salts-sucrose (TCBS) agar plates, which were incubated at 33 ºC for 24 h.
Bacteria of the genus Vibrio were identified at the species level according to the morphology of the colonies on TCBS agar (color, size, and shape), Gram staining, oxidase test, tolerance to NaCl concentrations (0%, 3%, 8%, and 14% of NaCl), and biochemical tests using the commercial API 20E test (bioMérieux, France).
Isolation of other bacteria
For the isolation of bacteria not belonging to the genus Vibrio, serial dilutions were inoculated on trypticase soy agar plates. Staphylococcus was identified by the morphology of the colonies and the presence of hemolysis in 5% blood agar, Gram staining, and coagulase and DNase enzymes.
Antibiotic sensitivity test
Resistance profiles for Vibrio were determined by the agar diffusion, or Kirby-Bauer, method according to the protocol established by the Clinical Laboratory Standards Institute (Uddin et al. 2018). Briefly, a bacterial suspension was prepared at a concentration of 0.5 McFarland scale, which was then inoculated on Mueller Hinton agar plates using a sterile swab. After 5 min, the discs were impregnated with antibiotics (tetracycline, ciprofloxacin, amoxicillin/clavulanic acid, ampicillin, and chloramphenicol) and placed on the agar surfaces. The plates were incubated at 37 ºC for 18-24 h; inhibition halos were measured; and results were recorded as resistant, intermediate, and sensitive (see Carpenter et al. 2018).
Statistical analysis
The Fisher test was used to compare the presence of bacteria considering the sampled communities and fish species. A one-way analysis of variance (ANOVA) was applied for bacterial loads, assuming normal distribution of the data after the Shapiro-Wilk test. All data were recorded and analyzed in SPSS v.21.
Results
Bacteria with yellow colonies were isolated on TCBS, with a diameter of 2-3 mm, Gram-negative bacilli, and negative growth at 0% (w/v) NaCl concentration and positive at 3%, 8%, and 14% NaCl concentrations. The oxidase, indole, citrate, hydrolysis of H2S, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, hydrolysis of urea, arabinose, and gas production tests were negative, whereas the motility, Voges-Proskauer, glucose, inositol, lactose, mannitol, sorbitol, and sucrose tests were positive; these reactions were compatible with those of Vibrio metschnikovii, which was found in 30.64% (95% CI: 18.36-42.92) of the analyzed fish samples. In addition, green colonies were isolated on TCBS, with a diameter of 2-3 mm, Gram-negative bacilli, and negative growth at 0% and 3% and positive at 8% and 14% NaCl concentrations. In this case, indole, citrate, hydrolysis of H2S, arginine dihydrolase, lysine decarboxylase, lactose, inositol, sorbitol, ornithine decarboxylase, hydrolysis of urea, arabinose, and gas production tests were negative, whereas the motility tests, Voges-Proskauer, glucose, mannitol, and sucrose tests were positive; these reactions were compatible with those of Salinivibrio costicola, which was found in 22.58% (95% CI: 11.37-33.79) of the analyzed fish samples. Gram-positive, coagulase-negative, catalase-positive, and DNase negative cocci bacteria were also isolated, and the characteristics were compatible with those of coagulase-negative staphylococci (CoNS) in 82.30% (95% CI: 71.94-92.57) of the fish samples.
Coinfections of V. metschnikovii, S. costicola, and CoNS were found in 5/62 samples; and S. costicola was exclusively found in 2/14 samples. CoNS was isolated in 33/52 samples (Fig. 2).
A significant difference (χ2, P = 0.032) was observed in the isolation of Vibrio spp. from the muscles of fish from the different communities. Frequencies of positive samples were higher for the Poneloya (5/8) and El Tránsito (8/13) communities and lower for the Las Peñitas (1/10) and Jiquilillo (10/31) communities (Fig. 1).
The positive percentage for V. metschnikovii was similar between the analyzed fish species (χ2, P ≥ 0.05), as the lowest percentage was found in C. latus (7.10%) and the highest in Pomadasys spp. (50.00%). Moreover, the positive percentage for S. costicola did not show significant differences between fish species (χ2, P ≥ 0.05), with frequencies ranging from 16.70% in L. colorado to 35.80% in M. cephalus. Isolation of CoNS was also similar between fish species (χ2, P ≥ 0.05), with frequencies ranging from 61.50% in M. cephalus to 94.40% in L. colorado.
The bacterial load for S. costicola was 1.58 × 103 CFU·g-1 in the analyzed fish samples from the Poneloya community. However, no significant difference was observed with respect to the load in fish samples from other communities (ANOVA, P = 0.075). The load of V. metschnikovii was 3.50 × 103 CFU·g-1 in samples from Las Peñitas and 1.57 × 103 CFU·g-1 in samples from Poneloya, whereas a load of 7.78 × 102 CFU·g-1 was found in samples from Jiquilillo and 5.5 × 102 CFU·g-1 in samples from El Tránsito (ANOVA, P = 0.024). The Vibro load in fish samples was 3.9 × 103 CFU·g-1 for Las Peñitas, 2.52 × 103 CFU·g-1 for Poneloya, 9.3 × 102 CFU·g-1 for Jiquilillo, and 5.25 × 102 CFU·g-1 for El Tránsito (ANOVA, P = 0.003), regardless of the bacterial species (Table 1).
Table 1 Bacterial load for the different Vibrio species in fishes from the studied communities.
| Vibrio species | Communities | N | Mean load (CFU·g-1) | Standard deviation | P value* |
| Salinivibrio costicola | El Tránsito | 4 | 2.25 × 102 | 1.89 × 102 | 0.075 |
| Jiquilillo | 4 | 5.75 × 102 | 4.57 × 102 | ||
| Las Peñitas | 1 | 4.00 × 102 | - | ||
| Poneloya | 5 | 1.58 × 103 | 1.03 × 103 | ||
| Vibrio metschnikovii | El Tránsito | 6 | 5.50 × 102 | 4.59 × 102 | 0.024 |
| Jiquilillo | 9 | 7.78 × 102 | 7.52 × 102 | ||
| Las Peñitas | 1 | 3.50 × 103 | - | ||
| Poneloya | 3 | 1.57 × 103 | 1.58 × 103 | ||
| Vibrio spp. | El Tránsito | 8 | 5.25 × 102 | 5.94 × 102 | 0.003 |
| Jiquilillo | 10 | 9.30 × 102 | 6.66 × 102 | ||
| Las Peñitas | 1 | 3.90 × 103 | - | ||
| Poneloya | 5 | 2.52 × 103 | 1.86 × 103 |
*According to the ANOVA test
Regarding macroscopic lesions, the presence of blisters in the operculum was observed in only 9.70% (6/62) of the fishes. Salinivibrio costicola was isolated from 4 of the 6 fishes with blisters and from 10 of the 56 fishes without blisters (χ2, P = 0.020). Vibrio metschnikovii was isolated from 2 of the 6 fishes with blisters and from 17 of the 56 fishes without blisters (χ2, P = 0.603). CoNS isolates were found in 82.30% (51/62) of total samples. They were found in 89.28% (50/56) of fishes without blisters and in 16.66% (1/6) of fishes with blisters, the latter being significantly lower than the former (χ2, P ˂ 0.01).
Regarding resistance to antibiotics, 100% (14/14) of S. costicola isolates showed resistance to amoxicillin/clavulanic acid but were sensitive to ciprofloxacin and chloramphenicol. On the other hand, 100% (19/19) of V. metschnikovii isolates were resistant to amoxicillin/clavulanic acid and sensitive to ciprofloxacin, and only 84.21% (16/19) showed sensitivity to chloramphenicol (Fig. 3).
Discussion
In this study, the Vibrionaceae species found in the fishes caught on the Pacific coast of Nicaragua were S. costicola and V. metschnikovii. The former is typical of marine environments, with few probabilities of causing diseases in humans due to ingestion; however, the latter species should be considered relevant when investigating foodborne illnesses associated with the consumption of raw or undercooked fish and shellfish (Matté et al. 2007). Vibrio metschnikovii has also been found in a variety of seafood (Nsofor et al. 2014) and it is considered a risk to human health, with virulence factors such as hemolysins and verotoxins (Austin 2010). In addition, there are reports of cases associated with wound infections, mainly in patients exposed to the marine environment, suggesting that V. metschnikovii could have zoonotic potential (Linde et al. 2004). A case of severe septic shock and cardiac arrest in a patient with V. metschnikovii was also reported (Jensen and Jellinge 2014). Vibriosis is a disease with worldwide spread in fish, affecting farmed fish under stress conditions (Martínez-Díaz and Anguas-Vélez 2002).
In this study, Vibrio was present in 38.71% of total samples, which is considered moderate when compared to other studies. For instance, a study isolating Vibrio spp. from mullets and tilapia in Venezuela obtained a total percentage of 38.9% for V. parahaemolitycus and 33.3% for V. cholerae (Arévalo et al. 2003). However, another study reported only 5.90% after isolating V. parahaemolyticus from 120 fish samples in Lima (Peru) (Aliaga et al. 2010).
In the present study the frequency of samples positive to Vibrio was higher for fish from the Poneloya community, although bacterial load was higher in samples from Las Peñitas and Poneloya. Both communities are close to the city of León, a place with high affluence of tourists; in addition, the fishing areas in both communities are close to estuaries, with the mouth of the Chiquito River in Las Peñitas and the mouth of the Telica River in Poneloya. In a study carried out in Chile, these factors were found to possibly influence the bacterial load in fish, as 80.00% of the river discharges in estuarine systems were contaminated as a result of improper handling of pharmaceutical, domestic, and industrial waste, a factor influencing the habitat and the spread of pathogens (Escobar 2002).
The analysis of macroscopic lesions showed that only 9.70% of fishes had blisters. This type of lesion has been associated with the presence and bacterial load of Vibrio spp. that can cause infections in commercially important aquatic organisms (Leyton and Riquelme 2008). In this study, no association was found between the presence of macroscopic lesions and the isolation of Vibrio. This could be related to the fact that, although many species belonging to the genus Vibrio are present throughout the marine environment and induce diseases in marine fish, disease outbreaks only occur when fish are stressed; therefore, this association is more likely to be observed in farmed fish than in those obtained from artisanal fishing (Martínez-Díaz and Anguas-Vélez 2002, Hashiem-Mohamed et al. 2016, Mahmoud et al. 2017).
The presence of CoNS in fish can be associated with anthropogenic contamination from the discharge of sewage high in fecal material into the estuaries, which distribute water to the open sea. It should be considered that the analyzed fishes were collected directly from the boats, so there was not much manipulation by the fishermen; that is, it is likely that the fishes were contaminated in their natural environment, as shown in a study carried out in Mexico, where authors detected Streptococcaceae, Micrococcaceae, and Staphylococus in freshly caught fish (Romero-Jarero and Negrete-Redondo 2011).
Regarding the antimicrobial resistance profiles in the genus Vibrio, resistance was high, mainly to amoxicillin/clavulanic acid. The way in which various antibiotics reach the oceans is presumably through the discharge of shrimp farm effluents into the oceans; although antibiotic use has been restricted because of the impact it has on the environment, antibiotics are still used to treat bacterial diseases, promoting the development of antibiotic resistant strains (Divyashree et al. 2020). A study on shrimp farming carried out in Mexico highlighted that the use of antibiotics has produced high resistance profiles in bacteria and promoted antibiotic persistence in aquatic environments (Santiago et al. 2009). In some regions, estuaries are highly productive and serve as important aquaculture production areas, and since the use of antibiotics is widespread in aquaculture, shellfish harvested from these areas are expected to be contaminated with bacteria harboring resistance genes (Kumar et al. 2005).
In conclusion, S. costicola, V. metschnikovii, and CoNS were isolated from fish from the Poneloya, Las Peñitas, Jiquilillo, and El Tránsito communities. Salinivibrio costicola is typical of marine environments and does not represent health risks, but V. metschnikovii causes diseases through the consumption of raw or undercooked fish. CoNS can also cause diseases and it is an indicator of anthropogenic contamination.











 texto en
texto en